Nuclear Aurora kinase A triggers programmed death-ligand 1-mediated immune suppression by activating MYC transcription in triple-negative breast cancer
Abstract
Background
Increasing studies have reported that oncogenes regulate components of the immune system, suggesting that this is a mechanism for tumorigenesis. Aurora kinase A (AURKA), a serine/threonine kinase, is involved in cell mitosis and is essential for tumor cell proliferation, metastasis, and drug resistance. However, the mechanism by which AURKA is involved in immune response regulation is unclear. Therefore, this study aimed to investigate the role of AURKA in immune regulation in triple-negative breast cancer (TNBC).
Methods
Peripheral blood mononuclear cells (PBMCs) were co-cultured with TNBC cells. The xCELLigence Real-Time Cell Analyzer-MP system was used to detect the killing efficiency of immune cells on TNBC cells. The expression of immune effector molecules was tested by quantitative real-time polymerase chain reaction (qRT-PCR) to evaluate immune function. Furthermore, to validate AURKA-regulated immune response in vivo, 4T1 murine breast cancer cell line with AURKA overexpression or downregulation was engrafted into BALB/c mice. The distribution and proportion of immune cells in tumors were further evaluated by immunohistochemistry and flow cytometry.
Results
Downregulation of AURKA in TNBC cells increased immune response by activating CD8+ T cell proliferation and activity. Nuclear rather than cytoplasmic AURKA-derived programmed death-ligand 1 (PD-L1) expression was independent of its kinase activity. Mechanistic investigations showed that nuclear AURKA increased PD-L1 expression via an MYC-dependent pathway. PD-L1 overexpression mostly reversed AURKA silencing-induced expression of immune effector molecules, including interleukin- (IL-2), interferon-γ (IFN-γ), and perforin. Moreover, AURKA expression was negatively correlated with the enrichment and activity of tumor-infiltrating CD8+ T cells in 4T1 engrafted BALB/c mouse model.
Conclusions
Nuclear AURKA elevated PD-L1 expression via an MYC-dependent pathway and contributed to immune evasion in TNBC. Therapies targeting nuclear AURKA may restore immune responses against tumors.
1 BACKGROUND
Human triple-negative breast cancer (TNBC), which lacks estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER-2), accounts for 15% of all breast cancers [1]. Compared to other subtypes of breast cancer, patients with TNBC typically have reduced overall survival, high malignancy, strong invasive ability, and high early recurrence rate [2, 3]. In addition, programmed death-ligand 1 (PD-L1) was found to be overexpressed in TNBC specimens and was associated with a high degree of malignancy and poor clinical prognosis [4]. The high expression of PD-L1 on the surface of TNBC cells inhibited T cell proliferation and promoted T cell apoptosis, forming immunosuppressive microenvironments [5-7]. Several clinical trials have used antibodies, such as pembrolizumab or atezolizumab, to block the PD-L1/programmed cell death protein 1 (PD-1) signaling and have revealed its objective response and effectiveness [8-11]. These studies have confirmed that PD-L1 can be used as an immunotherapy target for TNBC [12]. However, the mechanism by which PD-L1 expression is regulated in TNBC has not been fully elucidated.
Aurora kinase A (AURKA), a serine/threonine kinase, is involved in several stages of cell mitosis, including mitotic checkpoint regulation, microtubule stability, chromosome disjunction, and cytokinesis [13]. AURKA is also highly expressed in various types of tumors, such as gastric, colorectal, ovarian, liver, and breast cancers, promoting tumor development and progression [14]. AURKA also acts as an oncogene in breast cancer, and its expression is positively correlated with histological grade but negatively correlated with clinical prognosis [15]. Several studies have indicated that AURKA induces tumor cell proliferation, metastasis, and drug resistance in breast cancer by activating the downstream signaling pathways [16-18]. An early study from our group also revealed that AURKA was stained in the cytoplasm in the adjacent normal tissue, while it was highly detected in the nuclear fraction of breast cancer cells [19]. Nuclear AURKA interacts with MYC expression independently induced by heterogeneous nuclear ribonucleoprotein kinase, resulting in the enhancement of breast cancer stemness. However, the oncogenic function of nuclear AURKA has not been fully investigated.
Increasing studies have reported that oncogenes regulate components of the immune system, suggesting that this is a mechanism for tumorigenesis [20]. The gain-of-function of oncogenes suppressed anti-tumor immune response by regulating the expression of immune checkpoints. MYC enhanced PD-L1 and CD47 expression by directly binding to their promoters, and inactivation of MYC promoted anti-tumor immune response in TNBC [21]. Other oncogenes, such as mucin 1 (MUC1) and epidermal growth factor receptor (EGFR), have also been reported to induce PD-L1 expression [22, 23]. The mechanism by which AURKA regulates the immune response has not been fully determined. Recent work has shown that Alisertib (MLN8237), a selective kinase inhibitor of AURKA, increased the death of myeloid-derived suppressor cells (MDSCs) and suppressed the immunosuppressive functions of MDSCs, resulting in reduction of breast cancer progression, suggesting that AURKA contributes to anti-tumor immune responses [24].
Therefore, the purpose of the study was to investigate the role of AURKA in the regulation of immune response in TNBC.
2 MATERIALS AND METHODS
2.1 Reagents and antibodies
AURKA inhibitors VX-680 and MLN8237 and bromodomain and extra-terminal (BET) bromodomain inhibitor JQ1 were obtained from Selleck Chemicals (Houston, TX, USA) and dissolved in dimethyl sulfoxide (DMSO). MDA-MB-231 cells were treated with 0.1, 0.2, and 0.4 μmol/L VX-680 and 0.1, 0.2, 0.4, and 0.8 μmol/L MLN8237 for 24 h. MDA-MB-231 cells with NLS-AURKA were treated with 1 and 5 μmol/L JQ1 for 48 h for Western blotting detection. Antibodies against AURKA and phospho-AURKA (T288) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), goat anti-mouse IgG-horse radish peroxidase (HRP), and goat anti-rabbit IgG-HRP were obtained from Cell Signaling (Danvers, MA, USA). c-Myc was purchased from Santa Cruz Biotechnology (Dallas, TX, USA). PD-L1 was obtained from Abcam (Cambridge, MA, USA). TruStain FcX™ anti-mouse CD16/32, PerCP anti-mouse CD45, FITC anti-mouse CD3, PE anti-mouse CD4, APC anti-mouse CD8a, and PE anti-mouse CD69 used in flow cytometric analysis were purchased from Biolegend (San Diego, CA, USA). PE anti-human PD-L1 and its isotype control PE anti-human IgGk1 were obtained from BD Pharmingen (San Diego, CA, USA).
2.2 Patients
The present study was approved by the ethics committee of the Cancer Hospital of China Medical University, Shenyang, China (Ethics Review Approval no. 20170226). Between May 2017 and September 2018, a total of 60 patients with pathologically diagnosed TNBC who did not receive preoperative chemotherapy or radiotherapy were enrolled. Formalin-fixed paraffin-embedded tumor specimens were collected and were prepared in 5 μm tissue sections for immunohistochemistry (IHC) staining. Ethical practices were followed throughout to cover patient data confidentiality and compliance with the Declaration of Helsinki. All the participants signed informed consent. Clinical stage was classified according to the eighth edition of the American Joint Committee on Cancer TNM criteria for breast cancer [25].
2.3 UALCAN
UALCAN (http://ualcan.path.uab.edu), a comprehensive web resource, provided analysis based on RNA sequence and clinical data of 31 cancer types from The Cancer Genome Atlas (TCGA) database. Data of 719 breast cancer specimens (including 566 luminal subtype, 37 HER2-positive subtype, and 116 TNBC subtype) and 114 normal tissue specimens were obtained. AURKA expression was compared between the three breast cancer subtypes and normal tissues.
2.4 Public database KM-plotter analysis
Kaplan-Meier Plotter database (http://kmplot.com/analysis) was used to analyze prognostic values of AURKA expression in all breast cancers after the diagnosis.
2.5 Cell lines and mice
Human TNBC cell lines MDA-MB-231 and BT549 were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The murine mammary tumor cell line 4T1 was obtained from the American Type Culture Collection (Manassas, VA, USA). MDA-MB-231 cells were maintained in high-glucose Dulbecco's Modified Eagle's medium (Hyclone; GE Healthcare Life Sciences, Logan UT, USA). BT549 and 4T1 cells were cultured in RPMI-1640 (Hyclone). All medium was supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Cells were cultured in a 5% CO2 humidified incubator at 37°C. PD-L1 expression on BT549 and MDA-MB-231 cell surface was detected by flow cytometry. Six-week-old female BALB/c mice were purchased from Shanghai Laboratory Animal Center (Shanghai, China).
2.6 Plasmid construction and viral infection
Stable cell lines with overexpression or downregulation of AURKA expression were established and used in our previous study [19, 26]. Briefly, stable AURKA overexpression cell line was constructed with a lentivirus vector pLVX-AURKA. shRNA-mediated knockdown was performed with the pLKO.1 lentiviral vector containing specific shRNA against AURKA.
2.7 Cell viability assay
Cell viability assays were performed using the xCELLigence Real-Time Cell Analyzer (RTCA)-MP system (ACEA Biosciences, Inc., San Diego, CA, USA) as previously described [27]. Briefly, 1 × 104 cells were seeded into E-16 plates and placed in the incubator. After 6 h, peripheral blood mononuclear cells (PBMCs) isolated from healthy volunteers were co-cultured with parental MDA-MB-231 cells, shNC-transfected, or shAURKA-transfected MDA-MB-231 cells at the ratios of 1:4, 1:2, 1:1, 2:1, 4:1, 8:1, and 16:1. Tumor cells cultured without PBMCs were used as control. Cell viability was monitored every minute for 72 h. The cell index was normalized to the starting time of the co-culture.
2.8 Western blotting
Cells were harvested and washed with cold phosphate buffer saline (PBS). Total cell protein was extracted by using a Minute Total Protein Extraction Kit (Invent Biotechnologies, lnc, Minneapolis, MN, USA). Protein was quantified and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as previously described [28]. Primary antibodies were diluted and incubated overnight at 4°C. The membranes were then washed with Tris-buffered saline containing Tween 20 (TBST) and incubated with secondary antibodies at room temperature for 1 h. Chemiluminescence was detected using a BIO-RAD GelDoc XR+ system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The relative expression was quantified with ImageJ software (version 1.51 j8, National Institutes of Health and University of Wisconsin, Bethesda, MD, USA) using GAPDH as an internal control.
2.9 Quantitative real-time polymerase chain reaction (qRT-PCR)
qRT-PCR was performed as previously described [29]. Briefly, cells were lysed with 1 mL TRIzol reagent (Thermo Fisher Scientific, Carlsbad, CA, USA). cDNA was synthesized with PrimeScript™RT reagent Kit (TaKaRa, Shiga, Japan), then amplified by using SYBR Premix EXtaq (TaKaRa). PCR reactions were run using 2 μL of cDNA product by incubating for 30 s at 95°C, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. All samples were normalized to GAPDH. The primers for AURKA, PD-L1, GAPDH, interleukin (IL)-4, IL-10, transforming growth factor (TGF)-β, interferon-γ (IFN-γ), IL-2, MYC, granzyme B (GZMB), and perforin were as follows: AURKA: forward, 5’-GGAATATGCACCACTTGGAACA-3’, reverse, 5’-TAAGACAGGGCATTTGCCAAT-3’; PD-L1: forward, 5’-GGACAAGCAGTGACCATCAAG-3’, reverse, 5’-CCCAGAATTACCAAGTGAGTCCT-3’; GAPDH: forward, 5’-ACAACTTTGGTATCGTGGAAGG-3’, reverse, 5’-GCCATCACGCCACAGTTTC-3’; IL-4: forward, 5’-CGGCAACTTTGTCCACGGA-3’, reverse, 5’-TCTGTTACGGTCAACTCGGTG-3’; IL-10: forward, 5’-TCAAGGCGCATGTGAACTCC-3’, reverse, 5’-GATGTCAAACTCACTCATGGCT-3’; TGF-β: forward, 5’-CTAATGGTGGAAACCCACAACG-3’, reverse, 5’-TATCGCCAGGAATTGTTGCTG-3’; IFN-γ: forward, 5’-TCGGTAACTGACTTGAATGTCCA-3’, reverse, 5’-TCGCTTCCCTGTTTTAGCTGC-3’; IL-2: forward, 5’-TCCTGTCTTGCATTGCACTAAG-3’, reverse, 5’-CATCCTGGTGAGTTTGGGATTC-3’; MYC: forward, 5’-TCCCTCCACTCGGAAGGAC-3’, reverse, 5’-CTCGGTGCATTTTCGGTTGTTG-3’; GZMB: forward, 5’-TACCATTGAGTTGTGCGTGGG-3’, reverse, 5’-GCCATTGTTTCGTCCATAGGAGA-3’; perforin: forward, 5’-GACTGCCTGACTGTCGAGG-3’, reverse, 5’-TCCCGGTAGGTTTGGTGGAA-3’.
2.10 Mouse tumor model
Animal studies were approved and conducted according to the experimental animal guidelines of the China Medical University Animal Center. Six-week-old BALB/c mice were divided into three groups, with 6 mice in each group. Parental 4T1, AURKA-overexpressing 4T1, and shAURKA-4T1 cells (5 × 104) were inoculated into the ninth mammary fat pad. After 10 days, the tumor reached a measurable size. Tumor was measured every day since day 10 and quantified as 0.5 × length × width2. On day 24, the mice were euthanized, and tumor cells and splenocytes were prepared for flow cytometric analysis.
2.11 Flow cytometric analysis
PBMCs were co-culture with shNC- or shAURKA-transfected cells for 4 days. The proportions of T, natural killer (NK), and natural killer T (NKT) cells in PBMCs before and after co-culture were estimated by flow cytometry. Cultured cells (1 × 106) were harvested and suspended in 100 μL PBS. Cells were incubated with the indicated antibody at room temperature for 30 min. Mouse primary tumor cells were minced and disassociated with the EZ enzyme (Nitta Gelatin Inc., Osaka, Japan). Single cell suspensions from the spleen and mouse tumor sites were first incubated with TruStain FcX™ anti-mouse CD16/32 antibody to block non-specific staining and then stained on ice for 30 min with PerCP anti-mouse CD45, FITC anti-mouse CD3, PE anti-mouse CD4, APC anti-mouse CD8a, and PE anti-mouse CD69. Samples were detected by BD FACSCelesta™ (BD Biosciences, San Jose, CA, USA). Data were analyzed using FlowJo (BD Biosciences, Version 10). The cell surface expression of PD-L1 was expressed by Δ median fluorescence intensity (MFI) = MFI of PD-L1 - MFI of isotype control.
2.12 Carboxyfluorescein succinimidyl ester (CFSE) labeling
PBMCs were adjusted to a concentration of 1 × 106/mL, washed twice with PBS, and stained with 2.5 μmol/L CFSE at room temperature for 5 min. CFSE-labeled PBMCs were then washed with culture medium and co-cultured with tumor cells at a ratio of 5:1 for 96 h. CFSE staining was detected by BD FACSCelesta™.
2.13 IHC and evaluation
IHC staining was performed as previously described [30]. Briefly, slides were deparaffinized and rehydrated, and antigen retrieval was processed using Universal HIER antigen retrieval reagent (Abcam). Then, slides were treated with 3% H2O2 for 15 min to inhibit endogenous peroxidase activity and incubated with monoclonal anti-mouse AURKA antibody (A1231, 1:100; Sigma) and monoclonal anti-rabbit PD-L1 antibody (ab205921, 1:400; Abcam) overnight at 4°C. Slides were washed carefully and incubated with the secondary antibody. Rabbit-specific IHC polymer detection kit HRP/DAB (ab209101, Abcam) was used for the detection of PD-L1 staining. Dako REALTM EnVisionTM detection system (K500711, DAKO, Glostrup, Denmark) was used for AURKA. Slides incubated with non-immune serum IgG were used as negative control. The IHC staining score of AURKA was quantified by two independent experienced pathologists as described previously [18, 30]. A total score ≥5 was considered as high staining for AURKA. Tumor cell surface membrane staining > 5% was considered positive for PD-L1 [31].
Mouse tumor paraffin specimens were pretreated by using the above method. monoclonal anti-rabbit CD3 antibody (ab16669, 1:150; Abcam), monoclonal anti-rabbit CD11b (ab133357, 1:2000; Abcam), and monoclonal anti-rabbit Foxp3 (ab215206, 1:250; Abcam) were stained overnight at 4°C. Slides were washed and then incubated with goat anti-rabbit biotinylated secondary antibody (ab6720, 1:1000; Abcam) and visualized using an HRP-conjugated ABC system (ab7403, 1:10000; Abcam).
2.14 Statistical analysis
Statistical analysis was performed using the Statistical Package for the Social Sciences (SPSS) software version 21 (IBM Corp., Armonk, NY, USA). The Student's t-test was used to compare the participants’ age between the AURKA-low and -high groups. The chi-squared (χ2) test was used to compare other clinical characteristics between the two groups. Multiple comparisons between the groups were performed using Dunnett's least significant difference test. The PD-L1 IHC score was compared using the Fisher exact probability test. The data from 3 independent experiments are presented as mean ± standard deviation. P < 0.05 was considered statistically significant.
3 RESULTS
3.1 Downregulation of AURKA enhanced the cytotoxic activity of immune cells
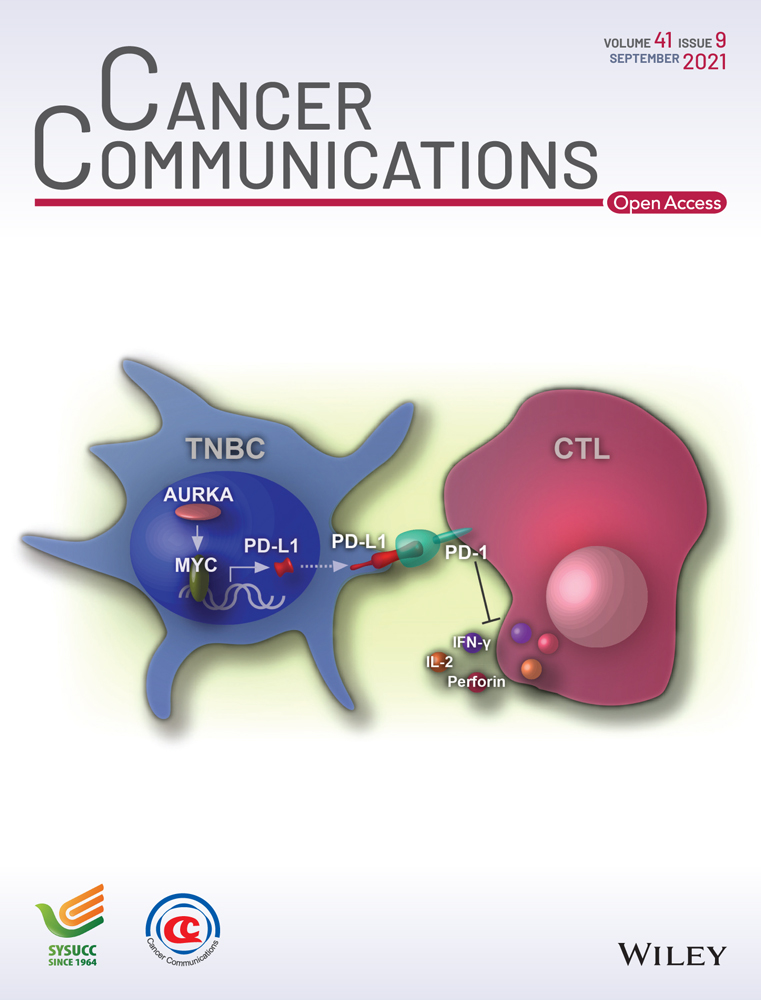
AURKA as an oncogene promotes the development of breast cancer [15]. The mRNA expression of AURKA was compared between the three breast cancer subtypes and normal tissues using data from the UALCAN database. As shown in Figure 1A, significantly increased expression of AURKA was found in breast cancer tissues, especially in TNBC. Further, using the Kaplan-Meier plotter database, we found that high expression of AURKA was significantly associated with poor prognosis after the diagnosis (Figure 1B). However, the mechanism by which AURKA affects the anti-cancer immune response is unknown. We first used the shRNA against AURKA and confirmed the downregulation of AURKA by qRT-PCR and Western blotting. AURKA expression was much lower in MDA-MB-231 cells transfected with shAURKA-1 and shAURKA-2 than in parental and shNC-transfected MDA-MB-231 cells (Figure 1C). To investigate whether AURKA expression inhibits immune cell cytotoxic activity, parental, shNC-transfected, and shAURKA-transfected MDA-MB-231 cells were co-cultured with PBMCs at various ratios, and cell viabilities were monitored using the RTCA-MP system every minute for 72 h. As presented in Figure 1D, tumor cell proliferation was inhibited by PBMCs in a dose-dependent manner. The viability of parental, shNC-transfected, or shAURKA-transfected MDA-MB-231 cells was inversely correlated with PBMC/tumor ratios (Pearson's correlation, r = −0.856, P < 0.01; r = −0.822, P < 0.01; and r = −0.641, P < 0.05, respectively). The cell viabilities of parental cells and shNC-transfected cells were not significantly different. However, MDA-MB-231 cells with downregulation of AURKA showed increased immune cell cytotoxic activity. Compared to parental or shNC-transfected cells, the viability of shAURKA-transfected cells was decreased at PBMC/tumor cell ratios of 1:2, 1:1, 2:1, 4:1, and 8:1 at 72 h. The ratio 4:1 was used in the following co-culture experiment. Tumor cells cultured without PBMCs served as control. After 72 h of co-culture, only a few shAURKA-transfected cells were attached to the dish (Figure 1E). As observed under the microscope, tumor cell death increased when exposed to PBMCs (Figure 1F). Compared to shNC, downregulation of AURKA enhanced target cell death. Additionally, after 72 h of co-culture, tumor cells and PBMCs were stained with CD45 and isolated by using a cell sorter. Cytokine expression in tumor cells and PBMCs was detected by qRT-PCR. Compared to shNC, downregulation of AURKA significantly inhibited IL-10 expression in tumor cells and increased IL-2 and IFN-γ expression in PBMCs (Figure 1G). These data indicated that knockdown of AURKA expression activated immune cell cytotoxic activity.
3.2 Downregulation of AURKA activated CD8+ T cell activity
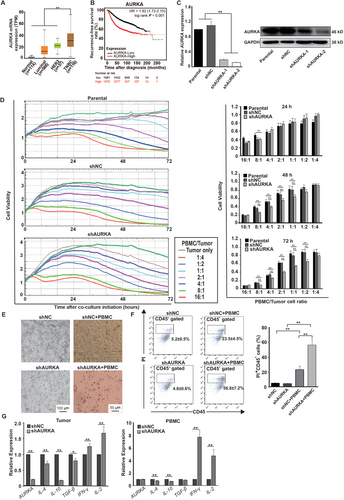
To investigate which effector cells were mainly affected by AURKA, we analyzed the proportions of T, NK and NKT cells in PBMCs before and 4 days after co-culture with shNC- or shAURKA-transfected cells. As shown in Figure 2A, compared to co-culture with shNC, knockdown of the expression of AURKA in MDA-MB-231 cells significantly increased the proportions of CD3+ T cells and CD3+CD8+ T cells, whereas the proportions of CD3+CD4+ T cells, CD3+CD56− NK cells, and CD3+CD56+ NKT cells were decreased. Next, we studied CD8+ T cell proliferation by analyzing the CFSE dilution on gate CD3+CD8+ T cells. CD8+ T cells exposed to shNC yielded a proliferating population of 32.2%, whereas CD8+ T cells exposed to shAURKA yielded a proliferating population of 69.3% (Figure 2B). Furthermore, we studied the effect of AURKA on cytolytic marker expression in CD8+ T cells. Co-cultured cells were collected and stained with CD107a on gate CD3+CD8+ T cells. As shown in Figure 2C, CD107a expression was increased significantly in CD8+ T cells when co-cultured with shAURKA-transfected MDA-MB-231 cells, but not increased in CD8+ T cells co-cultured with shNC-transfected cells. These data indicated that AURKA expression in MDA-MB-231 cells suppressed the proliferation and activation of CD8+ T cells.
3.3 AURKA induced PD-L1 expression
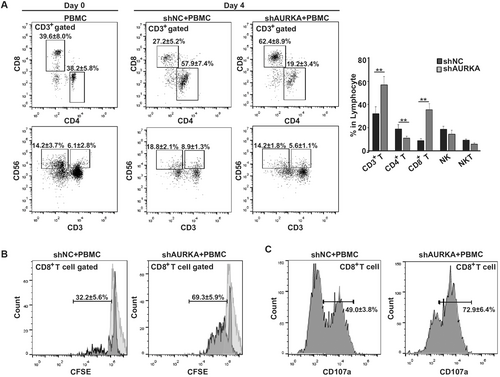
PD-L1 is highly expressed in TNBC and is associated with immune suppression, especially CD8+ T cells [4]. Several TNBC cell lines, such as BT549, MDA-MB-231, and SUM159 cells, have been reported to express PD-L1 [23]. Next, we investigated whether AURKA is involved in PD-L1 regulation. We used MDA-MB-231 cells stably overexpressing AURKA (AURKA), parental cells, and shAURKA-transfected cells to determine whether AURKA affects PD-L1 expression. Overexpression of AURKA resulted in enhanced PD-L1 expression, whereas AURKA knockdown resulted in PD-L1 downregulation at both the mRNA and protein levels (Figures 3A and 3B). Flow cytometry results supported that AURKA regulated cell surface expression of PD-L1 (Figure 3C). Similar data were obtained from another TNBC cell line, BT549 (Figure 3D). Since emerging evidence has shown that AURKA promoted tumor development through a kinase-independent mechanism [19, 32], we next tested PD-L1 regulation in the presence of the AURKA kinase inhibitor VX-680. AURKA kinase activity was inhibited, as p-AURKA expression was suppressed by VX-680 in a dose-dependent manner. However, the expression of PD-L1 along with AURKA in the test was comparable to that in the control group (Figure 3E). Flow cytometry data further confirmed that AURKA regulated PD-L1 expression on the cell surface in a kinase-independent mechanism (Figure 3F). Similar results were obtained by using another AURKA kinase inhibitor MLN8237 (Supplementary Figure S1). To further confirm that AURKA regulates PD-L1 in a kinase-independent manner, PD-L1 expression was compared between parental MDA-MB-231 cells and cells transfected with lentivirus expressing AURKA [WT or kinase-dead (KD)]. As shown in Figures 3G and 3H, PD-L1 expression was upregulated in both AURKA-WT and AURKA-KD cells at the mRNA and protein levels. Taken together, PD-L1 expression was not affected by AURKA kinase activity.
To verify the relationship between AURKA and PD-L1 expression in clinical samples, we performed IHC staining of AURKA and PD-L1 in 60 TNBC specimens. Clinicopathologic characteristics are shown in Table 1. AURKA was detected in the nucleus and cytoplasm in TNBC cells (Figure 3I). High nuclear expression of AURKA was observed in 35 (58.3%) samples, and PD-L1 positivity was detected in 12 (20.0%) samples. Furthermore, PD-L1 was only detected in the specimens with high nuclear AURKA expression (Figure 3I). Among the 48 PD-L1-negative samples, low nuclear expression of AURKA was found in 25 (52.1%) samples, whereas high AURKA expression was found in 23 (47.9%) samples. PD-L1 expression was significantly different between the nuclear AURKA low and high groups (P < 0.01).
| Nuclear AURKA expression | ||||
|---|---|---|---|---|
| Variable | Total | Low | High | P |
| Age [years; median (range)] | 53 (26-71) | 48 (32-71) | 54 (26-70) | 0.321 |
| Pathologic type [cases (%)] | 0.233 | |||
| Invasive ductal carcinoma | 59 (98.3) | 24 (40.0) | 35 (58.3) | |
| Others | 1 (1.7) | 1 (1.7) | 0 (0.0) | |
| Clinical stage [cases (%)]† | 0.017 | |||
| I | 8 (13.3) | 6 (10.0) | 2 (3.3) | |
| II | 38 (63.3) | 17 (28.3) | 21 (35.0) | |
| III | 14 (23.3) | 2 (3.3) | 12 (20.0) | |
| Lymph node metastasis [cases (%)] | < 0.001 | |||
| Negative | 36 (60.0) | 22 (36.7) | 14 (23.3) | |
| Positive | 24 (40.0) | 3 (5.0) | 21 (35.0) | |
| T stage [cases (%)] | 0.235 | |||
| T1 | 10 (16.7) | 6 (10.0) | 4 (6.7) | |
| T2 | 48 (80.0) | 19 (31.7) | 29 (48.3) | |
| T3 | 2 (3.3) | 0 (0.0) | 2 (3.3) | |
| T4 | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| N stage [cases (%)] | 0.003 | |||
| N0 | 36 (60.0) | 22 (36.7) | 14 (23.3) | |
| N1 | 10 (16.7) | 1 (1.7) | 9 (15.0) | |
| N2 | 12 (20.0) | 2 (3.3) | 10 (16.7) | |
| N3 | 2 (3.3) | 0 (0.0) | 2 (3.3) | |
| M stage [cases (%)] | ||||
| M0 | 60 (100) | 25 (41.7) | 35 (58.3) | |
| M1 | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
- † Clinical stage was classified according to the eighth edition of the American Joint Committee on Cancer TNM criteria for breast cancer
3.4 Nuclear AURKA induced PD-L1 expression
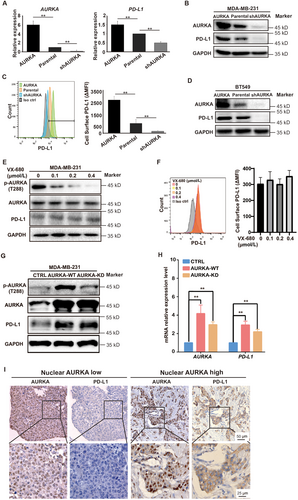
Our group previously demonstrated that nuclear localization of AURKA was required to enhance the stem cell phenotype of breast tumors [19]. Therefore, to investigate whether PD-L1 induction requires nuclear AURKA expression, we used the cell lines that we established previously. As shown in Figure 4A, AURKA was expressed in both the nucleus and cytoplasm of MDA-MB-231 cells. PD-L1 expression was enhanced in cells expressing nuclear AURKA but not cytoplasmic AURKA (Figure 4B), indicating that PD-L1 induction relied on nuclear AURKA expression. AURKA is associated with the upregulation of MYC in a variety of tumors including breast cancer [19], colon cancer [33], and ovarian cancer [34]. MYC has been shown to drive PD-L1 expression in TNBC [23]. However, whether AURKA-mediated PD-L1 induction depends on MYC signaling is unknown. We first used the MDA-MB-231 cells expressing nuclear AURKA or cytoplasmic AURKA and found that nuclear rather than cytoplasmic AURKA expression was associated with the induction of MYC and PD-L1 at both mRNA and protein levels (Figures 4C and 4D). Inhibiting nuclear AURKA expression in MDA-MB-231 cells by JQ1 suppressed MYC and PD-L1 expression (Figures 4E and 4F). To test if MYC acted as an important mediator in AURKA-promoted PD-L1 expression in TNBC, we knocked down MYC in both parental and AURKA-overexpressing MDA-MB-231 cells. As shown in Figures 4G and 4H, AURKA overexpression up-regulated PD-L1 in parental cells but not MYC-knockdown cells, suggesting that MYC is required for AURKA-promoted PD-L1 expression in TNBC. These data further support the hypothesis that nuclear AURKA induces PD-L1 expression via an MYC-dependent pathway.
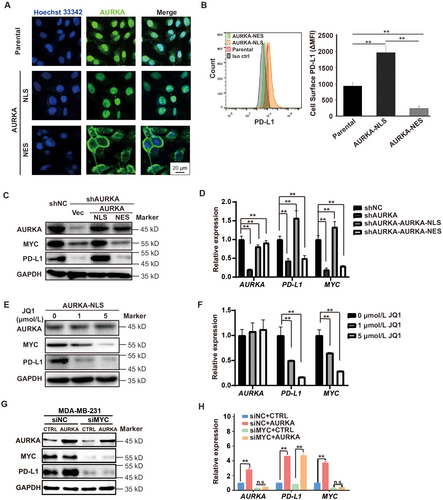
3.5 Downregulation of AURKA activated the immune microenvironment
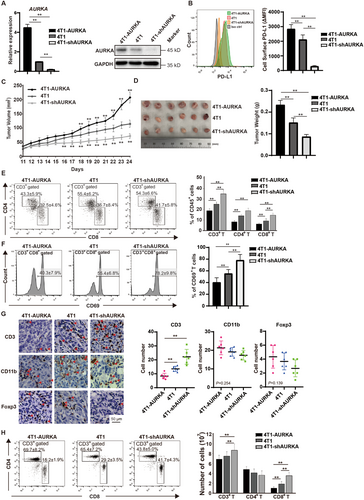
To analyze the alterations in the immune microenvironment after downregulation of AURKA, we established mouse 4T1 TNBC cell lines with AURKA overexpression (4T1-AURKA) or AURKA knockdown (4T1-shAURKA) and confirmed that AURKA induced PD-L1 expression by Western blotting and flow cytometry (Figures 5A and 5B). After transplanting into the mouse mammary fat pads, the growth of 4T1-AURKA xenograft was significantly enhanced, whereas downregulation of AURKA inhibited tumor growth (Figure 5C). The tumor weight of 4T1-AURKA xenografts was 1.71 times higher than that of 4T1 xenografts and was 2.43 times higher than that of 4T1-shAURKA xenografts (all P < 0.01) (Figure 5D). Analysis of the T cell population in the tumor on day 14 revealed that 4T1-shAURKA increased CD3+ T cells, CD4+ T cells, and CD8+ T cells (Figure 5E). Moreover, compared to the 4T1 group, the expression of CD69, an activation marker of CD8+ T cells, was markedly upregulated in the 4T1-shAURKA group (Figure 5F). Consistent with flow cytometry results, tumor-infiltrating T cells were increased in the 4T1-shAURKA group, whereas CD11b+ myeloid cells and Foxp3+ Tregs were slightly decreased (Figure 5G). Furthermore, we analyzed the absolute number and proportion of T cells in tumor-bearing mice and found that AURKA downregulation markedly increased CD8+ T cells in the spleen (Figure 5H). These data suggested that downregulation of AURKA stimulated T cell-mediated immune responses. To prove that PD-L1 acted as an important mediator in AURKA-promoted immunosuppression, we overexpressed PD-L1 in both MDA-MB-231-shNC and MDA-MB-231-shAURKA cells, Western blotting assays were conducted to evaluate the protein levels of AURKA and PD-L1 in tumor cells and the qRT-PCR were conducted to evaluate the mRNA levels of immune effector molecules (IL-2, IFN-γ, GZMB, perforin) in PBMCs. As shown in Supplementary Figure S2, depletion of AURKA attenuated PD-L1 expression, and activated PBMC functions, but overexpression of PD-L1 suppressed PBMCs function in both MDA-MB-231-shNC and MDA-MB-231-shAURKA cells. Taken together, these results demonstrate that PD-L1 plays an essential role in AURKA-mediated immunosuppression in TNBC.
4 DISCUSSION
In the present study, we demonstrated that TNBC cell lines with high AURKA expression decreased immune sensitivity by suppressing CD8+ T cell proliferation and activity. Moreover, PD-L1 was enhanced by the overexpression of AURKA and downregulated upon AURKA depletion by shRNA. However, PD-L1 expression was not repressed by the AURKA kinase inhibitor or kinase-dead mutant, which suggested the essential roles of nuclear AURKA.
Previous studies have shown that elevated nuclear AURKA was associated with poor clinical outcomes in breast cancer [19], ovarian cancer [35], and head and neck squamous cell carcinomas [36]. In the present study, we demonstrated that only nuclear AURKA induced MYC expression, and consistent with another study [23], MYC induced PD-L1 expression in TNBC. Moreover, inhibition of MYC with JQ1 or siRNA suppressed PD-L1 expression. These data suggest that PD-L1 induction by nuclear AURKA is dependent on MYC signaling. MYC is a key factor in tumorigenesis and maintenance. Recently, multiple groups have described that MYC regulated the expression of immune checkpoints, including CD47 and PD-L1 [21, 37]. Our group previously demonstrated that nuclear AURKA interacted with heterogeneous nuclear ribonucleoprotein (hnRNP) and activated MYC transcription by directly binding to the MYC promoter [19]. Hence, nuclear AURKA, as an MYC modulator, may have more functions in the immune response.
Tumor cell death might enhance immune destruction via eliciting the release of damage-associated molecular patterns (DAMPs) or pro-inflammatory cytokines [38], which indicated that AURKA silencing-caused PBMC activation might be partially due to AURKA silencing-induced apoptosis of cancer cells. IL-2, IFN-γ, and perforin expression was elevated in the AURKA-silenced cells but not in the AURKA-silenced cells with PD-L1 overexpression. These results indicated that PD-L1 was the major mediator in AURKA-induced immunosuppression (Figure 6).
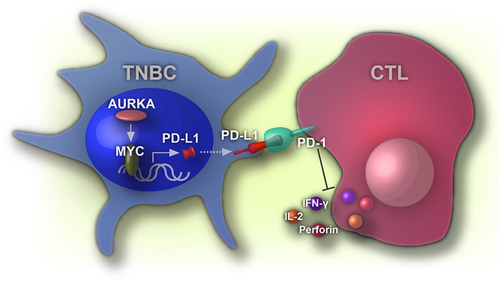
In the present study, we used murine breast cancer 4T1 cells to study the role of AURKA in immune response. As observed in human TNBC cells, AURKA induced PD-L1 expression in 4T1 cells. Downregulation of AURKA by shRNA activated CD8+ T cells. Moreover, AURKA downregulation not only enhanced the percentage of CD8+ T cell population in the tumor site but also elevated T cell abundance in tumor-bearing mice. The presence of tumor-infiltrating lymphocytes is associated with better clinical outcomes in patients with TNBC treated with chemotherapy [39, 40]. This may be another reason for AURKA overexpression contributing to chemotherapy resistance by regulating immune microenvironments. More recently, treatment of 4T1-bearing mice with an AURKA kinase inhibitor eliminated MDSCs and disrupted the immunosuppressive functions of MDSCs by inhibiting signal transducer and activator of transcription 3-mediated reactive oxygen species production [24]. These data suggest that AURKA regulates the tumor microenvironment in both myeloid and lymphoid cells by its kinase activity and its functions in the nucleus. Inhibition of both AURKA kinase activity and nuclear function may lead to strong immune evasion against tumors.
5 CONCLUSIONS
The present study showed that AURKA knockdown in tumor cells activated CD8+ T cell activity and that nuclear AURKA expression kinase-independently induced PD-L1 expression via activation of MYC signaling, resulting in immune evasion. In addition, downregulation of AURKA led to increased CD8+ T cell infiltration and activation in vivo. Therefore, therapies targeting nuclear AURKA may restore anti-tumor immune responses.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
All institutional and national guidelines for the care and use of laboratory animals were followed.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no conflict of interests.
ACKNOWLEDGEMENTS
The authors would like to thank Quentin Liu's lab members for technical support and critical comments. This research work was supported by the National Natural Science Foundation of China (No. 81702621 to SS, 81630005, 81820108024 to QL, 81972594 to MY, 82003141 to FP, No. 82002960 to BC, No. 31801100 to XD, No. 81703062 to LH), the National Key Research and Development Program, China (No. 2016YFC1303001 to HP), the Natural Science Foundation of Liaoning Province (20180550618 to SS, 2019-BS-081 to FP), the Guangdong Basic and Applied Basic Research Foundation (No. 2018A0303130299, 2020A1515010608 to MY), the “Seedling cultivation” program for young scientific and technological talents of Liaoning (No. LZ2020044 to FP, No. LZ2019067 to BC).
AUTHORS' CONTRIBUTIONS
HP, QL, and SS contributed to the conception and design of the study proposal. SS, WZ, XL, MY, XL, YL contributed to the methodology. SS and WZ performed data analysis. SS, WZ, QL, YZ, FA and FP wrote and revised the manuscript. HP and QL supervised the research process.