Targeting lactate dehydrogenase A (LDHA) exerts antileukemic effects on T-cell acute lymphoblastic leukemia
Abstract
Background
T-cell acute lymphoblastic leukemia (T-ALL) is an uncommon and aggressive subtype of acute lymphoblastic leukemia (ALL). In the serum of T-ALL patients, the activity of lactate dehydrogenase A (LDHA) is increased. We proposed that targeting LDHA may be a potential strategy to improve T-ALL outcomes. The current study was conducted to investigate the antileukemic effect of LDHA gene-targeting treatment on T-ALL and the underlying molecular mechanism.
Methods
Primary T-ALL cell lines Jurkat and DU528 were treated with the LDH inhibitor oxamate. MTT, colony formation, apoptosis, and cell cycle assays were performed to investigate the effects of oxamate on T-ALL cells. Quantitative real-time PCR (qPCR) and Western blotting analyses were applied to determine the related signaling pathways. A mitochondrial reactive oxygen species (ROS) assay was performed to evaluate ROS production after T-ALL cells were treated with oxamate. A T-ALL transgenic zebrafish model with LDHA gene knockdown was established using CRISPR/Cas9 gene-editing technology, and then TUNEL, Western blotting, and T-ALL tumor progression analyses were conducted to investigate the effects of LDHA gene knockdown on T-ALL transgenic zebrafish.
Results
Oxamate significantly inhibited proliferation and induced apoptosis of Jurkat and DU528 cells. It also arrested Jurkat and DU528 cells in G0/G1 phase and stimulated ROS production (all P < 0.001). Blocking LDHA significantly decreased the gene and protein expression of c-Myc, as well as the levels of phosphorylated serine/threonine kinase (AKT) and glycogen synthase kinase 3 beta (GSK-3β) in the phosphatidylinositol 3′-kinase (PI3K) signaling pathway. LDHA gene knockdown delayed disease progression and down-regulated c-Myc mRNA and protein expression in T-ALL transgenic zebrafish.
Conclusion
Targeting LDHA exerted an antileukemic effect on T-ALL, representing a potential strategy for T-ALL treatment.
1 BACKGROUND
T-cell acute lymphoblastic leukemia (T-ALL) is an uncommon and aggressive subtype of acute lymphoblastic leukemia (ALL), and it represents approximately 20% of adult and 10%-15% of childhood cases [1]. T-ALL prognosis has been significantly improved due to the widespread use of intensive chemotherapeutic protocols. However, approximately 20% of patients present with primary drug resistance, and the 5-year overall survival (OS) rate is approximately 50%; patients with relapsed disease still display an unfavorable outcome [2, 3]. Several clinical trials are currently exploring potential therapeutic targets, including the Notch-1 receptor 1 (NOTCH1) [4], the phosphatidylinositol 3-kinase (PI3K)/serine-threonine protein kinase (AKT)/mechanistic target of rapamycin kinase (mTOR) axis [5], the interleukin-7 receptor (IL7R)/Janus Kinase 1 (JAK)/signal transducer and activator of transcription (STAT) axis [6], cell cycle regulation [7], mitogen-activated protein kinase (MAPK) [8], the proteasome [9], and epigenetics[10], to improve the outcome of patients with relapsed disease [10]. The PI3K/AKT/GSK-3β signaling pathway is involved in the regulation of cell proliferation, differentiation, apoptosis, and glucose transport, and alterations in one or more components of this pathway have been frequently observed in ALL [11]. Studies have shown that in T-ALL patients with NOTCH1 mutations, the use of glutamine is the dominant source of intermediates for priming the tricarboxylic acid cycle (TCA) cycle, and combining Notch and glutaminolysis inhibitors is an effective treatment for mice bearing T-ALL primary grafts; thus, the therapeutic strategies focused on targeting glutaminolysis have been validated in this disease [12]. Furthermore, the PI3K/AKT signaling pathway has been reported to cause a metabolic switch from glutaminolysis to aerobic glycolysis in Notch-dependent T-ALL [12, 13], suggesting that targeting this metabolic pathway may be a potential strategy to improve T-ALL outcomes.
Regardless of oxygen availability, cancer cells prefer to use aerobic glycolysis for adenosine triphosphate (ATP) production; this is known as the Warburg effect [14]. Lactate dehydrogenase A (LDHA) is a key protein in the glycolytic pathway, which converts pyruvate to lactate. During this reaction, nicotinamide adenine dinucleotide (NAD+) is regenerated from (NAD)H in the absence of oxygen [15]. Serum lactic dehydrogenase (LDH) is an important prognostic factor predicting the clinical outcomes of both hematological and nonhematological malignancies [16, 17]. Serum LDH activity is increased in most patients with leukemia and lymphoma [18-20], and levels of this enzyme have prognostic value in both children and adults with lymphoma [21].
Oxamate is a derivative of pyruvate that inhibits the LDH-induced conversion of pyruvate to lactate, thus disrupting glycolysis [22]. Because cancer cells produce a large amount of energy via aerobic glycolysis, oxamate has been studied as an inhibitor of carbohydrate metabolism in various tumors [23-26]. In the study by Goldberg et al. [27], cells grown with low glucose or galactose levels produced very little lactic acid and were relatively insensitive to oxamate. As the property of aerobic glycolysis is unique to tumors rather than healthy mononuclear cells, oxamate might be slightly cytotoxic to healthy cells [27]. According to the Warburg effect, cancer cells prefer to obtain energy through the glycolytic pathway, and oxamate inhibits the key enzyme, LDH, of the glycolytic pathway. The antileukemic efficacy of oxamate is considered to be dependent on the proliferation rate of cancer cells [28].
We hypothesized that LDH may be involved in T-ALL progression and play an important role in the malignant behavior of T-ALL. To determine the role of LDH in the pathogenesis of T-ALL and the significance of LDHA in T-ALL progression and prognosis, we targeted LDHA to observe its effects on both primary T-ALL cells and T-ALL cell lines. We treated T-ALL cell lines with the LDHA inhibitor oxamate to investigate its potential antileukemic effects. CRISPR/Cas 9 gene-editing technology was applied to knock down LDHA and evaluate the effect of LDHA on T-ALL progression.
2 MATERIALS AND METHODS
2.1 Reagents and antibodies
Sodium oxamate, propidium iodide (PI), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), and all other chemical reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA). RPMI-1640 medium and fetal bovine serum (FBS) were obtained from Gibco/Thermo Fisher Scientific (Grand Island, NY, USA). The reactive oxygen species (ROS) inhibitor acetylcysteine (NAC) was purchased from Selleck (Houston, TX, USA). The following antibodies were used: anti-Bcl-2 (#2870), anti-AKT (#4691), anti-p-AKT (Ser473, #4060), anti-glycogen synthase kinase (GSK)-3α/β (#5676), anti-p-GSK-3α/β (#8566), anti-caspase-3 (#9665S), anti-caspase-9 (#7237S), anti-c-Myc (#5605), and anti-β-actin (#3700) purchased from Cell Signaling Technologies (Boston, MA, USA); anti-LDHA (AV54777) from Sigma-Aldrich; and horseradish peroxidase (HRP)-conjugated anti-mouse (#7076) and anti-rabbit IgG (#7074) from Kirkegaard & Perry Laboratories (Gaithersburg, MD, USA).
2.2 Cell culture
Jurkat cells were purchased from the American Tissue Culture Collection (ATCC) (Manassas, VA, USA), and DU528 cells were a kind gift from the A. Thomas Look Laboratory of the Dana-Farber Cancer Institute at Harvard Medical School (Boston, MA, USA). Jurkat and DU528 cells were cultured in RPMI-1640 supplemented with 10% heat-inactivated FBS and penicillin (100 U/mL)/streptomycin (100 μg/mL) and maintained at 37°C in a humidified incubator containing 5% CO2. Cells were passaged every 2 to 3 days and maintained at a density of 1-10 × 105/mL.
2.3 Patient samples
The study protocol was approved by the ethics committee of the Second Xiangya Hospital, Central South University, Changsha, China. Written informed consent was obtained from patients in accordance with the Declaration of Helsinki. Bone marrow samples of 27 patients with T-ALL diagnosed between 2016 and 2019 according to The 2008 World Health Organization (WHO) classification of lymphoid neoplasms were collected. Peripheral blood mononuclear cells (PBMCs) of 6 patients (3 males and 3 females; median age of 28.5 years, range of 20-66 years) and 6 healthy donors (3 males and 3 females; median age of 33.5 years, range of 22-68 years) were used for Western blotting. PBMCs of 3 patients (1 female and 2 males) and 3 healthy donors (1 female and 2 males) were used for the LDH activity assay. The diagnosis of T-ALL was based on morphology, cytochemical staining, and immunotyping according to the WHO classification of hematopoietic and lymphoid tissue tumor, the 4th edition.
2.4 Fish husbandry
Adult zebrafish (2-18 months old, weighted 0.55 ± 0.12 g) and embryos were kept at the Institute of Molecular Hematology of the Second Xiangya Hospital, Central South University, under a photoperiod of 14 h light and 10 h dark (lights on at 08:30, lights off at 22:30) in water at a pH of 7.2-7.6 and a temperature of approximately 28.5°C and were fed twice daily with newly hatched brine shrimp. Wild-type zebrafish were obtained from the Affiliated Hospital of Huazhong University of Science and Technology (Wuhan, Hubei, China); Hsp70-Cre and rag2-loxPdsRED2-loxP-EGFP-mMYC transgenic zebrafish were a generous gift from the A. Thomas Look Laboratory. Transgenic zebrafish expressing both Cre controlled by the heat-shock promoter and a rag2 promoter-regulated lox-dsRED2-lox-EGFP mMyc cassette rag2-LDL-EMyc in developing T cells was first generated by the A. Thomas Look Laboratory [29, 30]. The onset and progression of T-cell malignancy can be monitored by fluorescence microscopy as previously described [30, 31]. Wild-type zebrafish (6 females and 13 males) and transgenic zebrafish (5 females and 7 males) were used for the disease progression observation. Approximately 500 embryos were mainly used for injection of LDHA gRNA and Cas9 mRNA and observation.
2.5 Zebrafish with CRISPR/Cas9 knockdown of the LDHA gene
Guide RNA (gRNA) (5′-TAGGAGGCTATGGACTTGCAGCA-3′) was designed to target the LDHA protein-coding region at the exon 2 of zebrafish LDHA gene and selected with an online CRISPR Design Tool (http://crispr.mit.edu/). An LDHA-specific oligonucleotide was designed containing a 20-bp T7 promoter sequence (5′-TAATACGACTCACTATA-3′), the target sequence (5′-TAGGAGGCTATGGACTTGCAGCA-3′), and a 20-nt sequence that overlapped with a generic small guide RNA (sgRNA) template (5′-GTTTTAGAGCTAGAAATAGC-3′). This targeting oligonucleotide was annealed (conditions: 98°C for 2 min, 50°C for 10 min, and 72°C for 10 min) with a chimeric sgRNA core sequence (5′-AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC-3′) using Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific, Waltham, MA, USA). The product was reversely transcribed with the Transcript Aid T7 High Yield Transcription Kit (Thermo Fisher Scientific) to obtain sgRNA. Cas9 cDNA was synthesized using pT3TS-nCas9 (Plasmid #46757, Addgene, Watertown, MA, USA) as a template. The resulting cDNA product was purified using the Gene JET PCR Purification Kit (Thermo Fisher Scientific), and Cas9 mRNA was synthesized using the mMESSAGE mMACHINER Kit (T3Kits, AM1348, Life Technologies, Carlsbad, CA, USA). Approximately 300 one-cell-stage embryos were coinjected with purified LDHA gRNA and Cas9 mRNA using the MEGAclean™ Transcription Clean-Up Kit (Thermo Fisher Scientific). Each embryo was injected with 1 μL of a solution containing 300 ng/μL Cas9 mRNA and 200 ng/μL LDHA gRNA. Genomic DNA was extracted from six 2-day postfertilization (dpf) embryos injected with LDHA gRNA and Cas9 mRNA to verify the presence of mutations and confirm the activity of the gRNA. LDHA mutants were genotyped by TA cloning and PCR amplification. The selected parental (F0) adult zebrafish were hybridized with HSP70-Cre model zebrafish to obtain the first generation of hybrid offspring (F1) zebrafish embryos. After 2-3 months, DNA was extracted from the tail of F1 zebrafish, and PCR amplification with primers targeting HSP70Cre (forward primer: CGGGCATTTACTTTATGTTGC, reverse primer: GAAACCATTTCCGGTTATTCAAC) and LDHA (forward primer: CATTGAAGCAAACAATAAGTAGAGG, reverse primer: TGGAGCACATAGCTCAGGTTT) was performed using KOD FX PCR enzyme (TOYOBO, Osaka, Japan). After the aforementioned F1 zebrafish were sexually matured, they were hybridized with Rag2-LDL-EGFP-MYC model zebrafish to obtain the second generation of hybrid offspring (F2) zebrafish. The tails of F2 zebrafish were cut at 3 months old to extract DNA, and PCR amplification with LDHA primers was performed using KOD FX PCR enzyme. The resulting products were commercially sequenced to identify mutant fish. The MYC;Cre;LDHA+/− fish (containing MYC and Cre elements with LDHA half knockdown) constituted the experimental group, while MYC;Cre;LDHA+/+ fish (containing MYC and Cre elements without LDHA knockdown) constituted the control group.
2.6 MTT assay
Cell viability was determined using the MTT assay. In brief, Jurkat and DU528 cells and PBMCs from 5 patients were seeded in 96-well plates at 5000 cells/well and treated with different doses of oxamate in the presence or absence of NAC for the indicated times. At the end of each set of experiments, 20 μL of 5 mg/mL MTT was added to each well, and the cells were incubated for 4 h. Subsequently, the supernatants were aspirated, and the formazan crystals in each well were dissolved with 100 μL DMSO. The absorbance at 490 nm was measured in each well using an enzyme-linked immunosorbent assay reader (Thermo Fisher Scientific). The relative inhibition ratio was calculated as (1 - absorbance of oxamate-treated cells / absorbance of control cells) × 100%.
2.7 Colony formation assay
Jurkat cells and DU528 cells were exposed to various doses of oxamate for 24 h. Then, the cells were counted and plated in triplicate into 35-mm dishes in stemMACS methylcellulose medium (Miltenyi Biotec, Auburn, CA, USA) at 500 cells/mL. Colonies were counted under an inverted microscope after 12 days of culture (Olympus, BX 50, 400×, Tokyo, Japan). A colony was defined as a cluster of more than 50 cells.
2.8 Apoptosis assay
Apoptotic cells were quantified with an Annexin V-FITC Apoptosis Detection Kit (FACSCalibur, BD Biosciences, San Jose, CA, USA) according to the manufacturer's instructions. In brief, after incubating with various doses of oxamate for 48 h, Jurkat and DU528 cells were harvested, washed, suspended in binding buffer, and labeled with Annexin V-FITC and PI. Subsequently, the stained cells were analyzed by flow cytometry (FACSCalibur) and FlowJo LLC, Version 7.6.1 (BD Becton, Dickinson & Company, San Jose, CA, USA).
2.9 Cell cycle assay
Jurkat and DU528 cells were treated with various doses of oxamate for 48 h, after which they were harvested and fixed in 75% ethanol at -20°C. Next, the cells were washed, treated with 250 μg/mL RNase A, and stained with 50 μg/mL PI. The cell cycle distribution was analyzed using a flow cytometer.
2.10 Protein extraction and Western blotting
PBMCs were collected from T-ALL patients. T-ALL cell lines or cells collected from zebrafish embryos were raised until 3 days post-fertilization (dpf). The 3 dpf zebrafish were cut into two sections, half of which were used for PCR to verify whether LDHA was half knocked down. Zebrafish with LDHA half knockdown or not were collected and treated with RIPA lysis buffer (P0013B, Beyotime, Shanghai, China) on ice using ultrasonic grinding, and the protein concentration was measured using the BCA Protein Assay Kit (P0012, Beyotime). In all, 30 μg of total protein per lane was separated on dodecyl sulfate, sodium salt-polyacrylamide (SDS-PAGE) gels and then transferred onto polyvinylidene fluoride (PVDF) membranes, which were blocked in 5% nonfat milk and incubated overnight at 4°C with primary antibodies. The membranes were incubated with c-Myc (D84C12) rabbit mAb, β-actin (8H10D10) mouse mAb, caspase-3 (8G10) rabbit mAb, caspase-9 (D2D4) rabbit mAb, Bcl-2 (50E3) rabbit mAb, anti-LDHA antibody produced in rabbit, AKT (pan) (C67E7) rabbit mAb, p-AKT (Ser473) (D9E) XP® rabbit mAb, GSK-3α/β (D75D3) rabbit mAb, and p-GSK-3α/β (Ser21/9) (D17D2) rabbit mAb. Next, the membranes were treated with the appropriate HRP-linked anti-rabbit/mouse IgG secondary antibodies (Kirkegaard & Perry Laboratories) according to the species of the primary antibody. Protein signals were developed with an ECL detection kit (Pierce, Rockford, IL, USA). Protein bands from Western blotting were quantified by densitometry with Scion Image software (ImageJ 1.48u, Rawak Software Inc. Stuttgart, Germany).
2.11 LDH activity assay
Jurkat and DU528 cells were treated with various doses of oxamate for 48 h. The cells were centrifuged at 1500 rpm for 5 min, and the supernatant was collected. The LDH activity of the supernatant was measured using a Lactate Dehydrogenase Kit (Ningbo Meikang Biological Technology, Ningbo, Zhejiang, China) following the manufacturer's instructions. The LDH activity was read by a diagnostic automatic biochemical analyzer (Abbott, Lake Forest, IL, USA) at a wavelength of 340 nm.
2.12 LDH activity in PBMCs from T-ALL patients and healthy donors
Peripheral blood samples were collected from patients with T-ALL and healthy donors at the Second Xiangya Hospital, Central South University, China. The LDH activity in 3 patients (1 female and 2 males) and 3 healthy donors (1 female and 2 males) were assessed. PBMCs were lysed in extracting solution and disrupted by ultra-sonication. The LDH activity was then assessed on a diagnostic automatic biochemical analyzer (Abbott) at a wavelength of 340 nm.
2.13 Quantitative real-time PCR (qPCR)
For the analysis of gene expression in human T-ALL cell lines and zebrafish, total mRNA was extracted using RNAiso Plus (#9108, Takara Bio Inc., Kyoto, Japan), and purified mRNA was reversely transcribed into cDNA using the PrimeScript™ RT-PCR Kit with gDNAEraser (#RR014A, Takara Bio Inc.). The cDNA samples were then used for qPCR with the corresponding primers (C-MYC: 5′-TTCTCTCCGTCCTCGGATTC-3′, 5′-GTAGTTGTGCTGATGTGTGG-3′; β-actin: 5′-TTCCAGCCTTCCTTCCTGGG-3′, 5′-TTGCGCTCAGGAGGAGCAAT-3′). Real-time PCR was performed on a LightCycler 96 Real-Time PCR System (Roche, Basel, Switzerland) with SYBR®PremixExTaqII (#RR420, Takara Bio Inc.). With β-actin as an internal reference, the relative mRNA expression level of the target genes was calculated using the 2-△△Ct method.
2.14 Mitochondrial ROS assay
Jurkat and DU528 cells were cultured in 12-well plates at a density of 2 × 105/mL and treated with different concentrations of oxamate for 48 h. After treatment, the aspirated supernatant was incubated with MitoTrackerRedCM-H2XRos (#M7513, Thermo Fisher Scientific) at a concentration of 100 nmol/L at 37°C for 20 min, and mitochondrial ROS production was analyzed by flow cytometry.
2.15 TUNEL analysis of zebrafish cells
First, the F2 zebrafish were anesthetized using 150 mg/L Tricain MS222 (Sigma-Aldrich), placed on ice for 1 min to be killed, and dissected to obtain the kidneys. The kidneys were homogenized in phosphate-buffered saline (PBS) and then passed through a 40-mm filter to obtain a kidney cell suspension. Fish kidney cell smears were fixed with 4% paraformaldehyde in PBS and then blocked with 3% H2O2 in methanol. The slides were rinsed, incubated with the In Situ Cell Death Detection Kit (POD, Roche) according to the TUNEL staining protocol, and observed under an upright fluorescence microscope (ZEISS Scope.A1, Carl Zeiss, Jena Thueringen, Germany). Images were captured by a digital camera (AxioCamHrc, Carl Zeiss).
2.16 Assessment of tumor growth in zebrafish
The onset of leukemia was defined as a fluorochrome-labeled thymus that had grown to at least twice its normal size and that showed local infiltration without widespread dissemination, and T-ALL was defined as tumor cells that had widely disseminated throughout at least half of the fish [30].
2.17 Statistical analysis
The data were analyzed using SPSS 18.0 software (IBM, Armonk, NY, USA), and the graphs were generated using GraphPad Prism 5 software (GraphPad Inc., La Jolla, CA, USA). An unpaired two-tailed Student's t-test and ANOVA were used to compare the mean values. The results are presented as the means ± standard error of the mean, and P values ≤ 0.05 were considered significant.
3 RESULTS
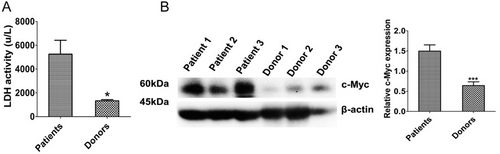
3.1 LDH activity and c-Myc expression in T-ALL patients
Clinical characteristics of the 27 T-ALL patients and 6 healthy donors are summarized in Table 1. The LDH activity of PBMCs was higher in the 27 T-ALL patients than in healthy donors (Figure 1A). c-Myc expression was increased in the patients’ T-ALL cells (Figure 1B). In our preliminary experiment, we observed that compared with PBMCs from healthy donors, Jurkat and DU528 cells displayed increased expression levels of both LDHA and MYC mRNA (data not shown). While stratifying the T-ALL patients by the age of under 18 years, 18-60 years, and over 60 years old, no significant differences in LDH activity were observed among these groups (all P > 0.05, data not shown).
| Characteristic | T-ALL patients | Healthy donors | P value |
|---|---|---|---|
| Total (cases) | 27 | 6 | / |
| Sex [cases (%)] | / | ||
| Female | 7 (25.9) | 3 (50.0) | / |
| Male | 20 (74.1) | 3 (50.0) | / |
| Age (years; mean ± SEM) | 30.22 ± 3.93 | 33.50 ± 3.55 | / |
| LDH activity (u/L; mean ± SEM) | 5251.74 ± 1164.00 | 1332.80 ± 99.80 | 0.004 |
| WBC (109/L; mean ± SEM) | 90.35 ± 21.27 | 7.37 ± 0.97 | 0.026 |
| Hb (g/L; mean ± SEM) | 105.19 ± 5.79 | 130.70 ± 4.26 | 0.027 |
| Platelet (×109/L; mean ± SEM) | 83.74 ± 18.07 | 189.30 ± 21.43 | 0.013 |
| Ratio of lymphoblast or pro-lymphocyte (%) | 70.0 ± 3.6 | / |
- Abbreviations: T-ALL, T-cell acute lymphoblastic leukemia; SEM, standard error of the mean; LDH, lactate dehydrogenase; WBC, white blood cells; Hb, hemoglobin.
3.2 Antiproliferative effects of oxamate in T-ALL cells
When Jurkat and DU528 cells were treated with oxamate at different concentrations (0, 1.25, 2.5, 5, 10, and 20 mmol/L) for 24, 48, and 72 h, oxamate exerted obvious proliferation-inhibiting effects (Figure 2A). The half-inhibitory concentration (IC50) values were 7.381, 3.753, and 3.197 mmol/L at 24, 48, and 72 h, respectively, in Jurkat cells and were 19.93, 7.418, and 5.016 mmol/L, respectively, in DU528 cells. The antiproliferative activity of oxamate was further confirmed by colony formation assays (Figure 2B). The MTT assay results revealed that when PBMCs from T-ALL patients were treated with oxamate for 48 h, there was significant inhibition of T-ALL cell proliferation, with an IC50 value of 18.36 mmol/L (Figure 2C).
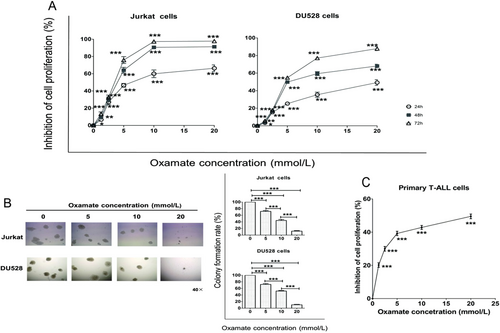
Oxamate inhibits the proliferation of Jurkat, DU528, and primary T-ALL cells. (A) Effects of different concentrations of oxamate (0, 1.25, 2.5, 5, 10, and 20 mmol/L) on Jurkat or DU528 cell proliferation after 24, 48, or 72 h of treatment. (B) Jurkat cells or DU528 cells were treated with the indicated concentrations of oxamate for 24 h and cultured for 14 days to observe the formation of colonies. The histogram shows the colony formation rates. (C) Effects of 48-hour oxamate treatment on the proliferation of primary T-ALL cells evaluated by the MTT assay. *P < 0.05, **P < 0.01, ***P < 0.001. All bars are presented as mean ± SEM.
Abbreviations: T-ALL, T-cell acute lymphoblastic leukemia; SEM, standard error of the mean; MTT, 3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-di- phenytetrazoliumromide
3.3 Effects of oxamate on cell cycle progression and apoptosis of T-ALL cell lines
We examined the effect of oxamate on the cell cycle progression of Jurkat and DU528 cells. Oxamate arrested both cell lines at the G0/G1 phase (Figure 3A). To further investigate whether oxamate induces apoptosis, we measured the apoptosis rates in cells treated with 1.25, 2.5, and 5 mmol/L oxamate for 48 h. As shown in Figure 3B, the apoptosis rates of both cell lines were significantly increased along with the increase of oxamate concentration (all P < 0.05).
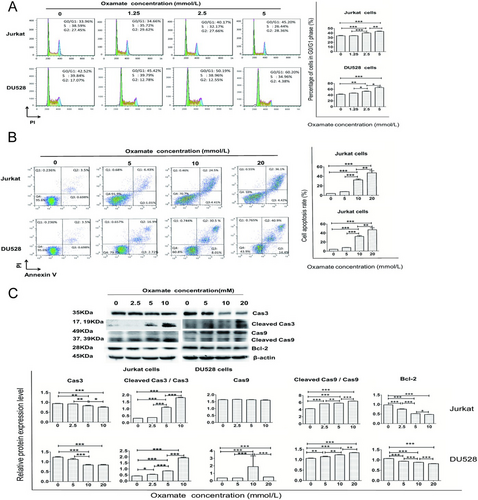
Oxamate induces T-ALL cell apoptosis and cell cycle arrest. (A) The flow cytometry results of Jurkat or DU528 cells after treatment with different concentrations of oxamate are shown. The number of cells arrested at the G0/G1 phase account for the majority of the population. (B) Jurkat and DU528 cells were treated with the indicated concentrations of oxamate for 48 h, and the percentage of apoptotic cells as tested by flow cytometry is increased with increasing concentrations of oxamate. (C) The expression of apoptosis-related proteins in Jurkat cells and DU528 cells after treated with oxamate for 48 h was measured using Western blotting. *P < 0.05, **P < 0.01, ***P < 0.001.
Abbreviation: T-ALL, T-cell acute lymphoblastic leukemia
We further examined apoptosis-related molecular markers in Jurkat and DU528 cells. As shown in Figure 3C, caspase-3 and caspase-9 were cleaved and activated following oxamate treatment. We also observed that oxamate down-regulated Bcl-2 expression in both cell lines, suggesting that oxamate-induced T-ALL cell apoptosis is associated with the activation of the intrinsic apoptosis pathway.
3.4 The effects of oxamate on regulating LDH activity
Jurkat and DU528 cells were treated with different doses of oxamate. The LDH activity was inhibited after oxamate treatment (Figure 4).
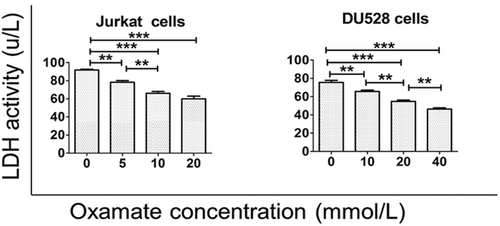
Oxamate increases the LDH activity in T-ALL cell lines. The LDH activity in Jurkat and DU528 cells was decreased with high concentrations of oxamate. *P < 0.05, **P < 0.01, ***P < 0.001.
Abbreviations: LDH, lactate dehydrogenase; T-ALL, T-cell acute lymphoblastic leukemia
3.5 The effects of oxamate on c-Myc expression in T-ALL cells
To elucidate the relationship between LDHA and c-Myc, we treated DU528 and Jurkat cells with different concentrations of oxamate. The data showed that oxamate down-regulated the mRNA and protein expression levels of both c-Myc and LDHA in both T-ALL cell lines (Figure 5).
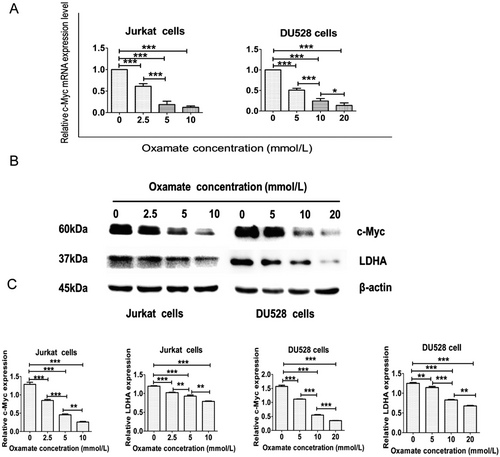
Oxamate down-regulates c-Myc expression in T-ALL cell lines. (A) The histogram shows significantly decreased c-Myc mRNA expression in Jurkat and DU528 cells after oxamate treatment. (B) Western blotting analysis of proteins extracted from Jurkat and DU528 cells using specified antibodies targeting c-Myc, LDHA, and β-actin. Cells were treated with increasing doses of oxamate for 48 h. (C) Relative protein levels of c-Myc and LDHA were further standardized to the level of β-actin in Jurkat and DU528 cells. *P < 0.05, **P < 0.01, ***P < 0.001.|.
Abbreviations: T-ALL, T-cell acute lymphoblastic leukemia; LDHA, lactate dehydrogenase A
3.6 The effects of targeting LDHA on the PI3K/AKT/GSK-3β signaling pathway
In the present study, we detected the protein levels of AKT, p-AKT, GSK-3β, and p-GSK-3β by Western blotting. The results revealed that oxamate down-regulated the levels of p-AKT and p-GSK-3β, while the expression levels of AKT and GSK-3β remained unchanged (Figure 6).
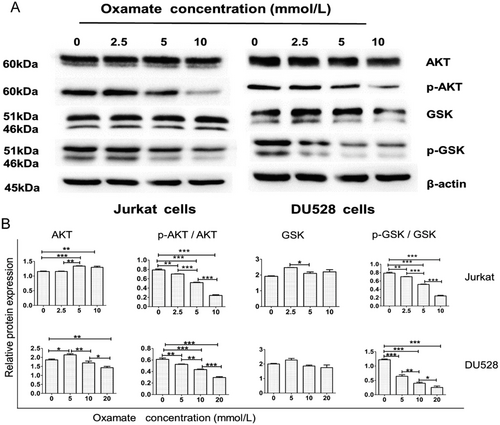
Targeting LDHA suppresses the PI3K/AKT/GSK signaling pathway. (A) Western blotting results for Jurkat and DU528 cells treated with different doses of oxamate for 48 h. (B) The relative protein levels of AKT and GSK and the ratios of p-AKT (serine/threonine kinase)/AKT and p-GSK/GSK were further standardized to the levels of β-actin in Jurkat and DU528 cells treated with the indicated doses of oxamate. *P < 0.05, **P < 0.01, ***P < 0.001.
Abbreviations: LDHA, lactate dehydrogenase; PI3K/AKT/GSK, phosphatidylinositol 3′-kinase /serine/threonine kinase/ glycogen synthase kinase 3 beta
3.7 The effects of oxamate on mitochondrial ROS production and growth of the T-ALL cell lines
Jurkat and DU528 cells were incubated with different concentrations of oxamate for 48 h, and the mitochondrial ROS production was analyzed. The results indicated that oxamate enhanced the mitochondrial ROS levels in Jurkat and DU528 cells (Figure 7A). When Jurkat and DU528 cells were concurrently treated with both NAC (a ROS inhibitor) and oxamate, cell proliferation were rescued (Figure 7B).
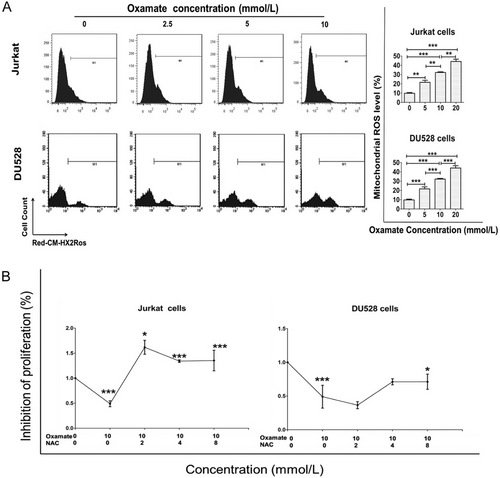
Oxamate enhances mitochondrial ROS expression in Jurkat and DU528 cells. (A) Jurkat and DU528 cells were treated with different concentrations of oxamate for 48 h, followed by staining with MitoTracker Red CM-H2XRos. Mitochondrial ROS production in cells was analyzed by flow cytometry. . (B) Treatment with NAC reversed the effect of oxamate in Jurkat and DU528 cells. The effects of oxamate and NAC on Jurkat and DU528 cell proliferation was evaluated by MTT assays. *P < 0.05, **P < 0.01, ***P < 0.001.
Abbreviations: ROS, reactive oxygen species. NAC, acetylcysteine
3.8 The effect of LDHA knockdown on T-ALL disease progression in transgenic zebrafish
To investigate whether LDHA affected T-ALL progression in zebrafish, we established a zebrafish model with LDHA knockdown by CRISPR/Cas9 technology, with MYC;Cre;LDHA+/+ zebrafish used as controls. The results showed that the onset of T-ALL was 7.3 days later in MYC;Cre;LDHA+/− zebrafish than in MYC;Cre;LDHA+/+ zebrafish (57.8 ±11.5 vs. 50.5 ±7.45 days, P = 0.042) (Figure 8A). We counted the fish that reached the stage of disease progression at weeks 5, 6, 8, and 20 since disease onset. The progression of tumors in MYC;Cre;LDHA+/− zebrafish group was significantly inhibited compared with that in MYC;Cre;LDHA+/+ zebrafish (P = 0.009) (Figure 8B), suggesting that knockdown of the LDHA gene in zebrafish can delay the progression of T-ALL. Interestingly, MYC;Cre;LDHA+/− zebrafish showed decreased expression of c-Myc in addition to LDHA (Figure 8C, D). As indicated by the results of TUNEL assays, the number of apoptotic leukemic cells was significantly increased in MYC;Cre;LDHA+/− zebrafish compared with MYC;Cre;LDHA+/+ zebrafish (Figure 8E), demonstrating that MYC;Cre;LDHA+/− zebrafish were sensitive to apoptosis.
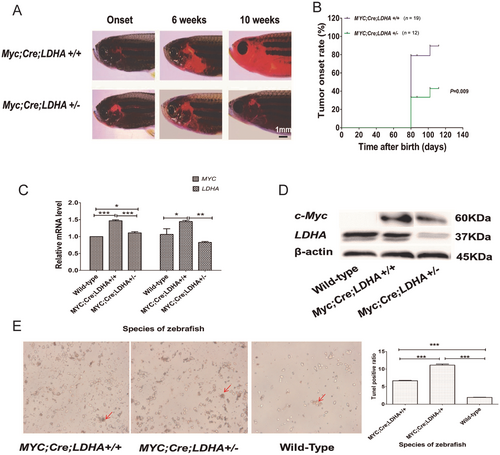
LDHA gene knockdown delays disease progression in MYC-induced T-ALL transgenic zebrafish. (A) Disease progression in MYC;Cre;LDHA+/+ and MYC;Cre;LDHA+/− zebrafish is shown in fluorescence images. (B) Tumor onset rate in MYC;Cre;LDHA+/+ and MYC;Cre;LDHA+/− zebrafish (P = 0.009). (C) The histogram shows significantly lower c-Myc and LDHA mRNA expression in MYC;Cre;LDHA-/+ zebrafish in MYC;Cre;LDHA+/+ zebrafish. (D) c-Myc and LDHA protein levels in MYC;Cre;LDHA+/+, MYC;Cre;LDHA+/−, and wild-type zebrafish. The relative LDHA level in MYC;Cre;LDHA+/− zebrafish was reduced by half compared with that in MYC;Cre;LDHA+/+ zebrafish. (E) TUNEL staining of apoptotic cells in MYC;Cre;LDHA+/+, MYC;Cre;LDHA+/−, and wild-type zebrafish. The red arrows indicate apoptotic cells. *P < 0.05, **P < 0.01, ***P < 0.001.
Abbreviations:LDHA, lactate dehydrogenase A; T-ALL, T-cell acute lymphoblastic leukemia
4 DISCUSSION
In the present study, we showed that targeting LDHA exerted anti-leukemic effects on T-ALL. First, we observed that the LDH activity in T-ALL patients was significantly increased compared with that in healthy donors. Based on clinical observations, we proposed that LDHA may be involved in the progression of T-ALL. Second, we confirmed that the LDH inhibitor oxamate suppressed proliferation and induced apoptosis in T-ALL cell lines and primary T-ALL cells. Third, we found that targeting LDHA could suppress the PI3K/AKT/GSK-3β signaling pathway and increase mitochondrial ROS levels in T-ALL cell lines. Finally, zebrafish models with LDHA knockdown and a predisposition to leukemia were established. We found that oxamate down-regulated c-Myc and resulted in delayed leukemic progression in MYC-induced transgenic zebrafish.
Oxamate is a derivative of pyruvate that inhibits the conversion of pyruvate to lactate via LDH, thus disrupting glycolysis [22]. Because cancer cells produce a large amount of energy via aerobic glycolysis, oxamate has been studied as an inhibitor of carbohydrate metabolism in various tumors [23-26]. LDH comprises four subunits, with the two most common subunits being LDH-M (encoded by the LDHA gene) and LDH-H (encoded by the LDHB gene, an important paralog of the LDHA gene). These two subunits can form possible five isoforms: LDH1 (4H), 2 (3H1M), 3 (2H2M), 4 (1H3M), and 5 (4 M). Read et al. [32] determined the crystal structures of two isoforms of human LDH—LDH-M and LDH-H. Both structures have been crystallized as ternary complexes in the presence of the NADH cofactor and oxamate. They found that the domain structure, subunit association, and active-site regions were indistinguishable between the two isoforms, suggesting that the distinctive activity of these enzyme isoforms is likely to result directly from the variation in charged surface residues peripheral to the active site [32]. Although oxamate is not an LDHA-specific inhibitor but rather targets all isoforms of LDH, our observations showed that oxamate could at least partially inhibit LDH activity.
Regarding whether oxamate can arrest cells in the G0/G1 phase, we did not analyze the cell cycle-related molecular markers CDK6 and cyclin D because these two markers for regulating the G1/S transition have been described in a previous study [33]. Indeed, if CDK6 and cyclin D expression were analyzed synchronously, the study would be more persuasive.
LDHA can convert pyruvate to lactate and couple with NAD+ cycling. LDHA was identified as a direct target gene of the c-Myc oncogenic transcription factor [34], but the connection between oncogenes and glycolysis is poorly understood [35, 36]. Tumor cells metabolize glucose to lactate even when oxygen is abundant, a phenomenon known as the Warburg effect [14, 15]. The Warburg effect has been directly linked to the activation of oncogenes, such as MYC, Ras, and AKT, which result in the deregulated conversion of glucose to lactate [37, 38]. Because hypoxia-inducible factor-1 (HIF), a key regulator of Warburg effect, also activates LDHA [34, 39], we hypothesized that a reduction in LDHA expression would inhibit cellular transformation and in vivo tumorigenesis because tumor tissues are hypoxic, and normal tissues neither are profoundly hypoxic or have activated MYC [36].
LDHA has been reported to be up-regulated in many cancer cells [40], and it favors tumor invasion and metastasis by promoting the metabolic switch to glycolysis [41]. LDHA is highly expressed in multiple myeloma cell lines, and targeting LDHA is considered a novel therapeutic approach [42]. These studies provide proof-of-concept that LDHA is a tractable therapeutic target.
In the present study, MYC inhibition by oxamate led to cell cycle blockade in the G1 phase. However, high levels of cleaved caspase 9 were observed in untreated Jurkat cells. The reasons may be as follows. First, cas9 cleavage is sensitive to the cell culture conditions. The untreated Jurkat cells showed slightly elevated levels of cleaved cas9, which may be partly related to the culture conditions. Second, when Jurkat cells were treated for Western blotting, the cells were serum-starved and lacked nutrients. Therefore, hunger-induced apoptosis is possible, explaining some of the background apoptosis. Third, Du et al. [43] observed caspase-dependent molecular mechanisms of anti-human DR5 monoclonal antibody mDRA-6 inducing apoptosis in Jurkat cells. Their data showed that the untreated Jurkat cells had 2.6% apoptotic cells detected using flow cytometry, which was similar to our observation (3.19%).
c-Myc primarily exerts its functions through transcriptional regulation of target genes that are associated with tumorigenesis and progression via different mechanisms. c-Myc promotes the Warburg effect, regulates LDHA expression in pancreatic cancer, and suppresses the in vivo conversion of pyruvate to lactate [44, 45]. Multiple genes in the glycolysis pathway are up-regulated in tumors, and their expression is rapidly repressed when c-Myc is knocked down [46]. c-Myc can also be regulated by targeting metabolic enzymes, including LDHA [47]. A previous study demonstrated that tumor progression occurred in an MYC-induced liver tumor animal model, and the LDHA activity was strongly associated with a lack of c-Myc activity [48]. The c-Myc-LDHA axis was reported to be closely associated with tumor progression, and it was indicative of a poor prognosis in pancreatic cancer patients [47]. c-Myc increases glucose uptake via the up-regulation of glucose transporters and regulation of lactate metabolism through the lactate transporter monocarboxylate transporter 1 (MCT1) and prevents pyruvate from entering the TCA cycle by modulating pyruvate dehydrogenase (PDK) expression [49]. Inhibiting c-Myc significantly decreases the Warburg effect. As LDHA is a key regulator of glycolysis, its inhibition can attenuate the growth of transplanted tumor cells [50, 51]; furthermore, the conversion from pyruvate to lactate was found to increase as tumors developed and was rapidly inhibited during regression [48].
In the current study, we observed that in both T-ALL cell lines and transgenic zebrafish, targeting LDHA could down-regulate c-Myc, which resulted in delayed leukemia disease progression in MYC-induced transgenic zebrafish. Interestingly, we found that metabolic changes precede not only tumor formation but also regression. Suppression of the c-Myc-LDHA axis contributes to the inhibition of tumor growth.
The repression of mitochondrial respiration leads to decreased ROS production and resistance to mitochondrial depolarization, thus reducing apoptosis [52]. Low to intermediate levels of ROS have been shown to stimulate cell proliferation and survival and induce the expression of stress response genes, whereas high levels of cellular ROS damaged DNA, promoted senescence, or initiated apoptosis [53, 54]. Recent studies have shown that attenuating LDHA activity in cancer cells using either small interfering or short-hairpin RNAs resulted in a shift to OXPHOS and decreased proliferation [50, 51, 55]. Knockdown of LDHA in breast cancer cells induced oxidative stress and apoptosis in vitro and in vivo [55]. Cell death in cancer cells with LDHA knockdown has been linked to increased oxidative stress activity [56]. LDHA-mediated inhibition of mitochondrial respiration and enhanced glycolysis in cancer cells resulted in decreased ROS production and resistance to apoptosis. In our observations, oxamate, an LDHA inhibitor, stimulated ROS production in the T-ALL cell lines.
The PI3K family is a family of closely related enzymes that are involved in a wide variety of cellular processes and expressed in various types of cancer. In normal and malignant lymphocytes, these enzymes play an important role in several signaling pathways. The key effector of AKT is mTOR, a serine/threonine kinase. AKT activation relies on the PI3K pathway, and it is recognized as a critical node in the pathway. This pathway is referred to as the PI3K-AKT-mTOR pathway [57]. Normally, GSK activity is inhibited by AKT activation, but the activity of both AKT and GSK can be down-regulated under certain conditions. Song et al. [58] demonstrated that a fusion protein NLS-RAR promoted both AKT and GSK-3β phosphorylation in NB4 cells (an acute promyelocytic leukemia cell line). The protein levels of both p-AKT and p-GSK-3β were decreased following pretreatment with the PI3K inhibitor LY294002 [58]. Jordan et al. [59] showed that myristoylated AKT blocked o-sulfated AGF function in a bladder cancer cell line, but up-regulated both p-AKT and p-GSK. Broecker-Preuss et al. [60] showed that the c-Myc and PI3K/AKT/mTOR pathways regulated glucose uptake in lymphoma cell lines and identified altered glucose metabolism as a potential target to improve inhibitor-based therapeutic approaches in these cells. After coincubation of the PI3K inhibitor LY294002 with rapamycin in lymphoma cells, they observed a decrease in LDHA expression, an increase in glucose-6-phosphate translocase (G6Ptase) expression, and decreased cell viability, which are consistent with our observations. These observations imply that AKT and GSK could be increased or decreased simultaneously.
In the current study, we mainly focused on the anti-T-ALL effects of targeting LDHA. Although LDHA is a key regulator of cell metabolism, we did not conduct metabolism-related experiments. Our preliminary observations suggested that the inhibition of T-ALL progression was through key regulators of tumorigenesis, namely, c-Myc and ROS. c-Myc increases the expression of many genes that support anabolic growth, including transporters and enzymes involved in glycolysis, fatty acid synthesis, glutaminolysis, serine metabolism, and mitochondrial metabolism [61]. The increased ROS production of cancer cells is due to a combination of oncogenic lesions and the tumor microenvironment. Hypoxia stimulates ROS production from the mitochondria and NOXs in cancer cells [62, 63]. Both c-Myc and ROS play important roles in regulating cancer metabolism [64]. Metabolism-related experiments should be conducted in the future studies. Regarding the samples from patients and healthy donors, we did not obtain sufficient samples of PBMCs to conduct MTT and LDH activity assays at the same time. Due to the current epidemic and time constraints, we are not able to conduct any other animal experiments related to the present study.
5 CONCLUSIONS
LDHA plays an important role in T-ALL progression, and targeting LDHA could inhibit T-ALL progression through the c-Myc-ROS and PI3K/AKT/GSK3β signaling pathways. Targeting LDHA may represent an alternative strategy for T-ALL treatment.
ACKNOWLEDGMENTS
We thank Jianfeng Zhou of Tongji Hospital Affiliated with Tongji Medical College for providing guidance and help with zebrafish knockdown technology (CRISPR/Cas9). We would also like to thank Yun Deng of Hunan Normal University for his assistance. Dr. A. Thomas Look of the Dana-Farber Cancer Institute at Harvard Medical School kindly provided the T-ALL transgenic zebrafish. This work was supported by the National Natural Science Foundation of China (81200368, 81670160) and the Hunan Natural Science Foundation (2017JJ2355).
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was performed in strict accordance with the recommendations of the regulations on human and animal experimentation of Central South University (No. 2012060). All experimental protocols were approved by the Ethics Committee of the Second Xiangya Hospital of Central South University (No. 2012298).
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
AUTHOR CONTRIBUTIONS
Haizhi Yu: Data analysis and cell line/fish line maintenance;
Yafei Yin: Experiments on T-ALL cell line performance;
Yifang Yi: Experiments on zebrafish model performance;
Zhao Cheng: Supervision of experiments and language editing;
Wenyong Kuang, Ruijuan Li, Haiying Zhong, Yajuan Cui, Lingli Yuan, Fanjie Gong, Zhihua Wang, Heng Li: Patient sample collection;
Hongling Peng: Project design and writing of the manuscript;
Guangsen Zhang: Project consultant and editing of the manuscript.




