Research highlights
A Redox Proteome ROSter
The accumulation of reactive oxygen species (ROS) within the cell can cause oxidative damage to vital cellular components such as proteins, lipids, and DNA, a condition referred to as oxidative stress. The thiol groups of cysteine residues in proteins are one of the most sensitive targets to ROS, rapidly oxidizing to form disulfide bonds with proximal cysteine residues, as well as higher oxidation states such as sulfinic and sulfonic acids. Although oxidative stress has been implicated in degenerative conditions such as aging and neurodegenerative disease, it is also a powerful antimicrobial defense initiated by mammalian cells to combat bacterial infection. As a protective measure, bacteria possess redox-sensitive proteins whose reversible oxidation enables processes such as regulation of gene expression and protein quality control to help the bacterium recover from the oxidative stress imposed by the mammalian host defense. Identifying the multitude of redox-sensitive proteins involved in the oxidative stress response would be an onerous and time-consuming task using traditional biochemical and genetic methods. However, in a recent report in PNAS, Lars Leichert and co-workers describe a method termed OxICAT—a thiol trapping technique combined with a mass spectrometric approach that can be used to quantitatively identify the target proteins of oxidation by hydrogen peroxide and hypochlorite and determine the redox-sensitive cysteines within the affected proteins. The authors applied their technique non-stressed and oxidatively stressed (H2O2 or NaOCl) E. coli to identify the redox-sensitive proteins. In so doing, they identified a number of proteins not previously shown to be redox-sensitive, roughly 30% of which are coded for by conditionally essential genes. The authors also found that the set of proteins with NaOCl-sensitive cysteines differed from those with H2O2 sensitivity, suggesting that these two physiological oxidants have different targets and perhaps different modes of action within the cell. Extending this technique to other cells and cell types may provide researchers with new antimicrobial targets or even lead to the development of better antioxidants and a better understanding of the link between oxidative stress and disease.
Leichert, L. I., et al. Proc. Natl. Acad. Sci. 2008 doi: 10.1073/pnas.0707723105.
Shaping Up the ER
Different organelles within the cell are characterized by their distinct shape, which is linked to their individual cellular function. What governs the shape of individual organelles, although integral to function, is oftentimes not well-understood. In the case of endoplasmic reticulum (ER)—a lacy, tubular network extending from the nucleus out into the cytoplasm that is involved in a variety of functions including protein translation, folding and transport—the tubular structure appears to be attributable to integral ER membrane proteins from two distinct protein families, according to a recent report in Science. Junjie Hu and co-workers showed that purified proteins from the yeast Yop1 family (human DP1) and the reticulon family were able to generate lipid bilayer tubules—using a variety of different lipids—when the purified proteins were reconstituted in proteoliposomes. Although unrelated sequentially, reticulon and Yop1(DP1) family proteins both contain a long hydrophobic central region, which was essential for tubule formation and which seems to form a hairpin turn that is inserted, wedge-like, into the membrane. The proteins also appear to be able to oligomerize to form arcs and rings. Other membrane-shaping proteins are known to utilize similar wedging mechanisms, and still other proteins have the ability to form ring or spiral scaffolds, but the combination of characteristics—as the authors propose for the reticulon and Yop1(DP1) proteins—could offer not only the ability to form narrow tubules with minimal surface coverage, but may also be a shaping mechanism employed by membrane proteins in other organelles.
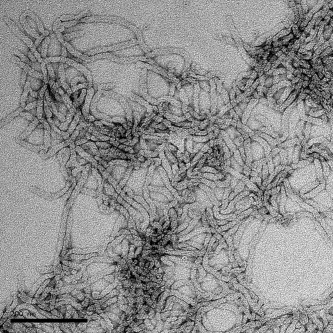
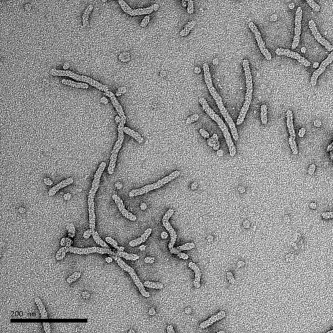
Hu, J., et al. Science 319, 2008, 1247–1250.




