Random Survival Forests With Competing Events: A Subdistribution-Based Imputation Approach
ABSTRACT
Random survival forests (RSF) can be applied to many time-to-event research questions and are particularly useful in situations where the relationship between the independent variables and the event of interest is rather complex. However, in many clinical settings, the occurrence of the event of interest is affected by competing events, which means that a patient can experience an outcome other than the event of interest. Neglecting the competing event (i.e., regarding competing events as censoring) will typically result in biased estimates of the cumulative incidence function (CIF). A popular approach for competing events is Fine and Gray's subdistribution hazard model, which directly estimates the CIF by fitting a single-event model defined on a subdistribution timescale. Here, we integrate concepts from the subdistribution hazard modeling approach into the RSF. We develop several imputation strategies that use weights as in a discrete-time subdistribution hazard model to impute censoring times in cases where a competing event is observed. Our simulations show that the CIF is well estimated if the imputation already takes place outside the forest on the overall dataset. Especially in settings with a low rate of the event of interest or a high censoring rate, competing events must not be neglected, that is, treated as censoring. When applied to a real-world epidemiological dataset on chronic kidney disease, the imputation approach resulted in highly plausible predictor–response relationships and CIF estimates of renal events.
1 Introduction
Survival analysis aims to model the time until the occurrence of a specific event (e.g., progression or death due to a certain disease) in dependence on a set of covariates. In clinical contexts, time-to-event data are often collected in observational studies that are prone to right censoring. Right censoring happens, for example, when patients drop out of a study or do not experience their event before the end of the observation period. In addition to a single event of interest, other event types are often recorded in observational studies and present in survival datasets. Often, the occurrence of these competing events cannot be assumed to be independent of the occurrence of the event of interest, especially if shared underlying (disease) mechanisms or shared risk factors are present.
An example would be examining kidney failure (KF) as the event of interest in patients with chronic kidney disease (CKD), while death by other causes than KF is a competing event (Hsu et al. 2017). In the German Chronic Kidney Disease (GCKD) study (Titze et al. 2015), for instance, 5217 participants with CKD are followed up annually, so data can be evaluated at the discrete time points corresponding to 1-year time intervals. One of the aims of the study is to better understand the factors underlying the progression of the disease. Potential risk factors that were collected in the study at baseline included, for example, leading kidney disease, as well as kidney function measures, such as serum creatinine, estimated glomerular filtration rate (eGFR), and U-albumin/creatinine ratio (UACR). Since CKD is a risk factor for heart failure (HF) and CKD and HF share common risk factors (Beck et al. 2015), death (e.g., from cardiovascular causes) should be considered as a competing event.
A popular approach to analyzing survival data (i.e., time to first event) in the presence of competing events is the subdistribution hazard model by Fine and Gray (1999), which extends the classical Cox proportional hazard model (Cox 1972). The Fine and Gray model introduces a subdistribution hazard function, which is a modification of the hazard function in traditional survival analysis. This function quantifies the instantaneous rate of the event of interest occurring, given that the subject has not yet experienced the event of interest until that time (assuming that the event of interest will never occur first once a competing event has already occurred (cf. Fine and Gray 1999).
As with other classical regression approaches, the subdistribution hazard model is not designed for high-dimensional data settings or complex covariate–risk relationships. In such scenarios, machine learning models such as deep survival neural networks (e.g., Giunchiglia, Nemchenko, and van der Schaar 2018; Gupta et al. 2019; Lee et al. 2018) and random survival forests (RSF) can be applied (Ishwaran et al. 2008; Schmid, Wright, and Ziegler 2016; Wright, Dankowski, and Ziegler 2017). While neural networks can be most beneficial for unstructured data, such as text and images, random forests might be advantageous for structured data exploration and for identifying important clinical covariates (Archer and Kimes 2008). Also, random forests are easy to train and require less resource-consuming hyperparameter tuning.
While numerous methods for competing events exist in classical regression, only some implementations of machine learning models for survival analysis consider competing events. In existing approaches, competing events in random forests are addressed by, for example, adapting the split rules (Ishwaran et al. 2014; Therrien and Cao 2022) or by using pseudo-value regression approaches (Mogensen and Gerds 2013). The latter method transforms the categorical event status into a continuous pseudo-value. Consequently, a random forest with regression trees is fitted instead of survival trees.
In this paper, we take a different approach for modeling competing risk data with RSF: Rather than introducing new split rules or new architectures for competing events, we transform the competing event problem into a single-event problem. This is achieved by manipulating the (input) dataset via an appropriately defined imputation scheme. More specifically, we consider three types of imputation approaches: In the first approach, the dataset is only preprocessed once before training the RSF. In the other two approaches, the dataset is adjusted directly at the tree instance of the forest: at the root node of the trees or at every node of the trees. As a consequence, well-established split rules and variable importance measures of single-event RSF can be applied. Also, the cumulative incidence function (CIF) for the event of interest can be directly calculated from the output of the single-event RSF.
The idea of using imputed censoring times instead of the observed competing event time has been applied successfully already for classical statistical modeling and neural networks: Ruan and Gray (2008) presented an imputation approach for continuous-time and semiparametric models based on Kaplan–Meier estimates. Gorgi Zadeh, Behning, and Schmid (2022) took a similar approach and proposed a method to train single-event deep neural survival networks on competing-event data, in which the unobserved censoring times of subjects with a competing event were imputed using subdistribution weights.
In this article, we describe the proposed methods and use a simulation study to evaluate their applicability and performance metrics in different situations. Finally, we report on a first application of the methods to real data obtained in the GCKD study.
2 Methods
2.1 Discrete Survival Analysis for Competing Risks
The aim of our proposed method is to estimate the CIF for an event of interest given a set of covariates. In a typical setting with right-censored data, we assume to follow-up the subjects with baseline covariates . Either an event time or a censoring time is observed, with the status indicator and the type of event denoted by . For each subject, either the event of interest (, ), a competing event (, ) or a censoring event () is observed. Just as in Fine and Gray's modeling approach, all competing events are combined into one single competing event, denoted . We assume that the event time and the censoring time are independent random variables (random censoring). In a naive approach, where competing events are ignored and treated as censored, the random censoring assumption may be violated. We further assume that the censoring mechanism is noninformative, meaning that the distributions of and do not share any common parameters. In our approach, time is modeled on a discrete scale (possibly after grouping the continuous times into intervals), that is, , where denotes the maximum observable time (interval). This is motivated by the observation that most versions of RSF implicitly treat time as an ordinal variable (Ishwaran et al. 2008), and that many other available implementations of machine learning methods also use discrete-time data structures (e.g., Ren et al. 2019).
In this article, we focus on modeling the occurrence of the event of interest (). The CIF for the event of interest is defined as , so the probability of experiencing the event of interest at time or prior with a given set of covariates .
2.2 Random Survival Forest for Single Events
The central architecture of the RSF is similar to the standard random forest approach (Breiman 2001; Ishwaran et al. 2008). In the first step, a number of (bootstrap) samples are generated. Next, a survival tree (Hothorn et al. 2004) is grown on each bootstrap sample. At each tree, covariates are considered for splitting into child nodes, and the best split is selected. Many split rules have been proposed, including splitting based on the maximum log-rank statistic, C-index, Hellinger distance, and many more (Schmid et al. 2020). In this paper, we use the log-rank statistic, which can deal with both continuous and discrete-time data. The tree is grown until it reaches a termination constraint, for example, tree depth, minimum number of observations, or if no increase with respect to splitting criteria is possible. A cumulative hazard function (CHF; ) is calculated at the terminal nodes of each tree. Averaging across all trees leads to the ensemble CHF. In settings without competing events, the CIF can be obtained from the CHF by .
2.3 Imputation Using Subdistribution Weights
- 1. Single imputation of the entire (training) dataset, performed outside the RSF architecture.
- 2. Imputation in the root node of each tree in the dataset. With this approach, the weights are calculated on the subset of data in the respective tree only.
- 3. Imputation in each node of each tree. Here, the weights are calculated only on the samples present in the respective node.
To gain an understanding of the distribution of the true compared to the imputed , or the resulting , for the three imputation approaches, please see the Illustration subsection below.
2.3.1 Implementation
We incorporated the described imputation approaches in the C++ implementation of the R package ranger (Wright and Ziegler 2017). The implementation involved adding a function to the survival trees that calculates a life table estimate of the censoring survival function analogous to the function estSurvCens of the R package discSurv (Welchowski et al. 2022). The C++ command line interface has been used for benchmarks described below. The source code can be found here https://github.com/cbehning/ranger.
2.4 Simulation Setup
We conducted a simulation study to investigate whether subdistribution-based imputation in the case of competing events can improve the estimation of the CIF in RSF compared to ignoring the competing events.
2.4.1 Data-Generating Mechanisms
In each simulation run, we created a set of subjects , with . For each subject, we first generated a vector of 50 normally distributed covariates . Next, three time variables were created: a time for the event of interest, a time for the competing event, and a censoring time . Afterwards, we sampled from a binary distribution with parameter whether the event of interest () or the competing event () was observed (see below). Next, the status indicator was generated as follows: the subject was censored if the censoring time was before the event time (). If the censoring time for this subject was after the event time, the subject remained uncensored ().
To discretize the continuous event times, we categorized the event times into intervals with interval borders obtained by empirical quantiles of width . The empirical quantiles were pre-estimated once per parameter from an independent sample with 1,000,000 observations.
The following two covariate–risk relationships were investigated in this simulation study.
2.4.2 Setup 1: Tree-Like Covariate–Risk Relationship
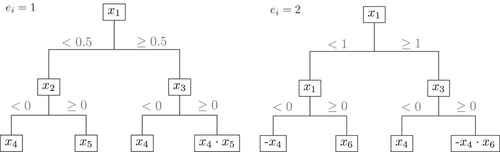
2.4.3 Setup 2: Interactions
In this setup, the covariates , and are associated with both events, while and are only associated with the event of interest and and are only associated with the competing event. Only the interaction term is shared between both linear predictors. Thus, the dependency structure is similar to Setup 1, but here is added.
- 1. Reference: If a subject experiences the competing event, this is replaced with the true (simulated) censoring time in the dataset. With these input data, the RSF for single events will model the true censoring rate and serves as a reference (see Table 2A).
- 2. Naive approach: Ignoring the competing event and treating the competing event time as if a censoring happened (see Table 2B).
- 3. Impute once (imputeOnce): Single imputed dataset before fitting the standard single-event RSF implementation (see Table 2C).
- 4. Impute in root (imputeRoot): RSF implementation with imputation in each root node, thus imputing once in each tree on all subjects available at the tree's root node (see Table 2D).
- 5. Impute in each node (imputeNode): RSF implementation with imputation in every node, thus imputing multiple times per tree on the subjects available in the respective node (see Table 2D).
| Time | Status | Event | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 15 | 1 | 1 | 15 | 18 | ||||
| 2 | 7 | 2 | 2 | 7 | 12 | 0.7352 | |||
| 3 | 13 | 0 | 1 | 17 | 13 | 0.3930 | 1.2740 | ||
| 4 | 10 | 2 | 2 | 10 | 20 | 0.0181 | |||
| 5 | 3 | 1 | 1 | 3 | 14 | 0.7355 | 1.3222 | ||
| A: Simulated | B: Naive approach | C: Impute once | D: Impute in forest | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Time | Status | Time | Status | Time | Status | Time | Status | ||||
| 1 | 15 | 1 | 1 | 15 | 1 | 1 | 15 | 1 | 1 | 15 | 1 |
| 2 | 12 | 0 | 2 | 7 | 0 | 2 | ? | 0 | 2 | ?? | 0 |
| 3 | 20 | 0 | 3 | 13 | 0 | 3 | 13 | 0 | 3 | 13 | 0 |
| 4 | 10 | 0 | 4 | 10 | 0 | 4 | ? | 0 | 4 | ?? | 0 |
| 5 | 3 | 1 | 5 | 3 | 1 | 5 | 3 | 1 | 5 | 3 | 1 |
In each simulation run, we divided the dataset into a training () and a test set () before applying the methods above. Splits were stratified by the event types (event of interest, competing event, censoring). We carried out 1000 simulation runs for each combination of setup, parameters and and for each imputation method, resulting in an overall number of 90,000 simulation runs. We chose 1000 runs because this number guaranteed the width of the reference limits for the CIF (provided in Figures S6 and S7) to be smaller than 0.1 (i.e., ). Apart from the described incorporated imputation approaches, the RSFs were fitted using the R package ranger from the command line interface with default parameters. This means fitting trees with covariates selected in each node (), the log-rank split rule, sampling with replacement, and a minimal node size of 3.
2.5 Illustration
To gain an understanding of the distribution of the imputed censoring times in the imputeOnce method compared to the true censoring times and the censoring times used in the naive approach (), Figure S3 depicts the distribution of and for one simulation run. As the censoring times are imputed multiple times in imputeNode and imputeRoot this visualization would be less meaningful, and we show the variation across life table estimates of instead. To illustrate the variability of the life table estimates across trees and nodes, Figures S4 and S5 show examples for a setup, a simulation run, and a combination of and . Here we see that the estimation of G on the subsets in the trees (imputeRoot) leads to increased variability of . The estimation in each node of the trees (imputeNode) increases the variability even further. The mean squared error between the true censoring time and the imputed censoring times was lowest for imputeOnce and highest for imputeNode (see captions of Figures S3–S5).
2.6 Performance Measures
2.6.1 Calibration Graph
The agreement between the reference and estimated CIFs was evaluated using calibration graphs. Here, we directly compared the estimated CIF (averaged over the 1000 simulation runs) across the different (imputation) methods on the test dataset. The method containing the simulated true censoring times instead of the imputed censoring times served as a visual reference (see Table 2A). Generally, the methods are well calibrated if the averaged estimated CIF curve agrees closely with the reference.
2.6.2 C-Index
The concordance index (C-index) was used to evaluate the discriminatory power of the different model fits on the test data. The C-index essentially measures how well the ranking of the (time-averaged) estimated CHFs matches the ranking of the observed event times. A stronger alignment between these rankings with higher C-index values implies greater discriminatory power. The C-index as implemented in the function cIndex in the R package discSurv (Welchowski et al. 2022; Heyard et al. 2020) was calculated.
2.6.3 Brier Score
The predictive performance of the approaches was compared using the Brier score (Gerds and Schumacher 2006). The Brier score at time point is defined as the (estimated) squared difference between the observed and modeled status () at that time. The integrated Brier score (IBS) is calculated by integrating the Brier score over all possible time points . Lower values imply a better prediction. The Brier score was calculated using the R package pec (Mogensen, Ishwaran, and Gerds 2012).
3 Results
The calibration graphs in Figure 2 (simulation Setup 1) and Figure 3 (simulation setup 2) show the CIF on the test dataset that was not seen during training, averaged over 1000 simulation runs. They include nine different scenarios, that is, nine combinations of the parameters and , where determines the rate of the event of interest, and affects the censoring rate.
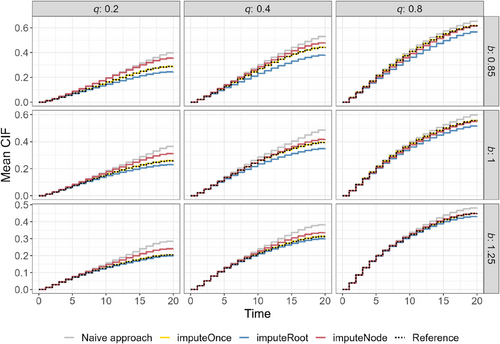
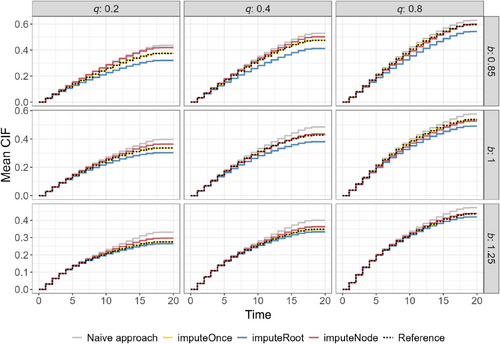
In all scenarios, all RSF architectures show similar CIF estimates for the first time points and tend to diverge for later time points. Here, the naive approach (gray lines), where competing events are treated as censoring, always shows the strongest overestimation and highest deviation from the reference method (dotted lines). The CIF of the imputeOnce approach visually overlaps with the dotted reference line that was obtained by training the single-event RSF on the simulated (true) censoring times (Reference).
The methods where the imputation is directly implemented in the nodes of the trees show the highest differences in the setting with a low censoring rate (, first row). In all settings, the method with only one imputation in each root node tends to underestimate the CIF. In contrast, the imputation in each node tends to overestimate the CIF, especially in the scenario that corresponds to a low event-of-interest rate and a low censoring rate (). For a better understanding of the overlap of the estimated CIF, Figures S6 and S7 provide reference limits for the estimated CIF at time points 10, 15, and 20.
Concerning the C-index and the Brier score, all methods perform similarly Tables S1–S4). The methods that do not impute directly in the random forest (imputeOnce, naive approach) performed slightly better with regard to these metrics in Setup 2. However, in Setup 1, the imputation in the root nodes of the RSF (imputeRoot) performed similarly to imputeOnce. To further gain insight on the properties of the simulation design, we divided the 1000 simulation runs into 10 batches. Using these batches, an estimate of the Monte Carlo error was calculated. The corresponding results are presented in Figures S8–S15.
In addition to the described performance measures, we calculated the permutation variable importance (VIMP) on the training dataset using the time-aggregated CHF as a marker in Harrell's C-index (cf. Ishwaran et al. 2008). Figures S16–S20 show the 10 variables with the highest mean permutation VIMP averaged over 1000 simulation runs of the training datasets in Setup 1. Note that the covariates to were included in the data-generating mechanism for the event of interest (only the covariates and were associated on a continuous level), while the covariates were associated with the competing event (see Figure 1). The variables and are indeed the two most important variables throughout for the reference, the naive approach, and imputeOnce, while mostly only was considered in the first 10 variables for imputeRoot and imputeNode in the scenarios with a low and medium rate of the event of interest (). In scenarios with a high rate of the event of interest (, were included for imputeRoot and imputeNode.
For Setup 2 (Figures S21–S25), the variables associated with the event of interest to are among the five most important variables in all scenarios for the Reference, the naive approach, and imputeOnce. In contrast, for the approaches imputeRoot and imputeNode, variables that are not associated with the event of interest get selected, especially in the scenarios with lower . For imputeRoot and imputeNode, all of the variables to are only included in the first most important variables when the censoring rate is high (). For the high censoring scenario, the variables and , which are associated with the competing event, are also in the top 10 most important variables.
3.1 Limitations
We acknowledge that our simulation study has several limitations: First, our study did not have a preregistered study protocol. This was mainly because we designed our simulation study to gain insight into the properties of the proposed methodology and to provide the first empirical evidence on its functioning (“phase II” in the framework by Heinze et al. 2024). Clearly, more extended simulations covering a broader range of scenarios (corresponding to later phases in the framework by Heinze et al. 2024) will have to be based on preregistered protocols. Second, our simulation study used a rather limited set of values for the parameters , and . We chose these values because they had already been used in previous simulation studies with competing events (Beyersmann, Allignol, and Schumacher 2011; Berger et al. 2020), thus making our design consistent with earlier publications. Third, simulation Setups 1 and 2 were chosen to represent data-generating mechanisms with multiple interactions and arbitrary cut-offs, allowing us to mimic a scenario in which we typically would not fit a classical Cox proportional hazards model. These setups could be extended by data-generating mechanisms in which the competing event and the event of interest do not share risk factors (not explored in our simulations). They could further be extended to high-dimensional data settings where the number of covariates exceeds the number of observations.
4 Application to the GCKD Study Data
- Demographic: age (in years), sex (male/female), alcohol (low-normal drinking/heavy drinking), smoking (nonsmokers/former smokers/smokers), family status (single/married or in a stable partnership/separated or divorced/widowed), number of siblings, number of people living in the household, employment (fully employed/part time/ housework/pension/job-seeker/training/other), private insurance (yes/no), professional qualification (still in training/apprenticeship/master (craftsperson)/university degree/without degree/other/unknown);
- Clinical: enrollment (inclusion based on low eGFR value or proteinuria), body mass index (BMI, in ), hypertension (yes/no), coronary heart disease (CHD: yes/no), stroke (yes/no), asthma (yes/no), chronic obstructive bronchitis (COPD: yes/no), taking painkillers (regularly/when required/never/unknown);
- Laboratory: serum creatinine (in ), eGFR (in ), UACR (in ), CRP (in ), low-density lipoprotein (LDL) cholesterol (in ), high-density lipoproetin (HDL) cholesterol (in );
- Family history: number of siblings with stroke, number of siblings with kidney disease.
We compare the approaches described above on a complete case subset of the GCKD dataset. The dataset included 4256 participants. Of those, 412 () reached KF (event of interest), and 409 () died without reaching KF first (competing event, participants who died due to forgoing dialysis or transplantation are considered as KF). The estimated CIF and the 10 covariates with the highest VIMP can be seen in Figure 4. The CIF is lowest for the imputeOnce approach and imputeRoot. Due to the high sample size, imputeRoot and imputeOnce may lead to similar imputation results. We suspect the CIF of these two approaches to be the most realistic estimate based on the results of the simulation study, where the naive approach and imputeNode generally overestimated the CIF. Although the differences appear small, they will presumably become even more relevant with the longer observation period that can be evaluated in the future. The imputation method proposed by Ruan and Gray (2008) included analyses of multiple imputed datasets instead of a single imputation. Therefore, we performed 10 imputations of the imputeOnce method and compared the pooled results to single runs (see Table S11). In this application, however, the variability of the estimated CIF was quite low.
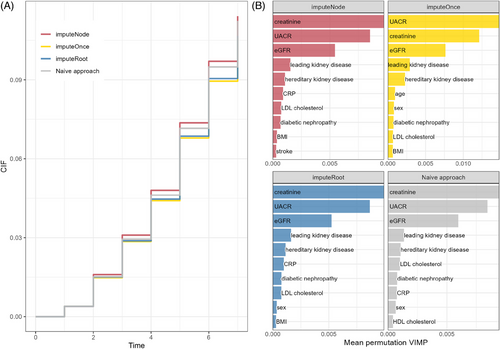
All approaches describe creatinine, UACR, eGFR, the leading CKD cause, and having a hereditary disease cause as the first five most important variables. This is followed by CRP, LDL cholesterol, and having diabetic nephropathy for imputeNode, imputeRoot, and naive approach. For imputeOnce, the demographic parameters age and sex were selected next instead of the laboratory parameters. The order of the selected covariates differs slightly between the approaches. A table containing VIMP values for all four methods can be found in Table S12. Both eGFR and UACR are reasonable covariates, as their progression is being discussed as a surrogate endpoint for progression to KF (Levey et al. 2020).
5 Conclusion
We have proposed three variants of a subdistribution-based imputation approach to handle competing risks in RSF. Our simulation study showed that the CIF is well estimated when imputation already takes place outside the forest on the training data (imputeOnce).
In survival analysis, the occurrence of competing events must be appropriately taken into account. The naive approach of considering competing events as censoring can lead to biased estimates of the CIF, although our simulation study has shown that this approach may lead to similar results in terms of C-index and IBS. Differences in the estimated CIF became apparent, especially in scenarios with a high censoring rate or a low rate of the event of interest. By including the naive approach in the simulation, we wanted to raise awareness for the proper treatment of competing events when using machine learning applications.
It should be emphasized that the naive approach estimates the cause-specific CHF of the event of interest, ignoring the hazards of the competing events. Hence, it cannot be directly transformed into the event of interest's CIF. While the CIF for the event of interest could be derived from a combination of all cause-specific hazard functions, we chose not to use this approach due to its complexity in analyzing covariate effects. Instead, we preferred the Fine and Gray method, as it provides a single (direct) effect per covariate. With random forests and other machine learning methods, having such a direct effect per covariate is a major advantage, in particular when it comes to the interpretation of measures like variable importance. Furthermore, the Fine and Gray method can reduce the computational effort, as it avoids having to fit separate machine learning models (one per cause-specific hazard). Also, note that the performance of the cause-specific hazard approach may strongly depend on the availability of sufficient numbers of observed events in the data.
A major finding of our simulation study is that imputing the estimated censoring times once before fitting the random forest (imputeOnce) essentially results in unbiased CIF estimates. Compared to imputations of the estimated censoring times in every tree node (imputeNode) or in the root node of the trees (imputeRoot), imputeOnce showed a systematically better performance with respect to the calibration graph of the CIF.
- i. In contrast to single imputation, with imputeNode, the sample sizes for estimating are much smaller, especially in the direction of the terminal nodes, which are usually very small for RSF (default minimum node size: 3 in our simulation study). Therefore, the estimation of weights is less accurate, probably translating into less accurate, or even biased, estimates of CIF. In the imputeRoot scenario, the sample size is smaller than that of imputeOnce for subsamples, while with bootstrapping, there are additional problems due to ties, which can also lead to biases in the CIF estimates. We have seen this in the GCKD data: With a large sample size and a higher number of events, the differences between imputeOnce and imputeRoot are smaller and the estimate of stabilizes.
- ii. With imputeNode, the censoring survival function is reestimated in each node and thus on the subset of data that is available in the specific node. Consequently, due to smaller sample sizes in the lower levels of the trees, imputeNode tends to show much higher variability in the estimation of the censoring survival function than imputeOnce. The censoring times might thus be imputed with reduced precision, resulting in a decreased estimation accuracy of the CIF. Similar arguments hold for the imputeRoot strategy (effectively operating on data samples with a reduced size).
In conclusion, the proposed single-imputation strategy (imputeOnce) allows for converting the competing-risks setting into a single-event setting. All RSF features and options (split rules, variable importance measure, etc.) are immediately available for this setting, making it much more straightforward to apply RSF in the competing-risks context. Issues for future research include a comparison to other machine learning methods and other techniques for dealing with competing events in RSF. This could, for example, be done in the framework of a neutral comparison study (see, e.g., the recently published Special Collection on “Neutral Comparison Studies in Methodological Research” in Vol. 66 of Biometrical Journal).
Acknowledgments
M.N.W. was supported by the German Research Foundation (DFG) - Emmy Noether Grant 437611051. The German Chronic Kidney Disease (GCKD) study was funded by grants from the German Federal Ministry of Education and Research (BMBF, grant number 01ER0804), the KfH Foundation for Preventive Medicine, and corporate sponsors (http://www.gckd.org). We are grateful for the willingness of the participants to participate in the GCKD study. The enormous effort of the study personnel of the various regional centers is highly appreciated. We thank the large number of nephrologists who provide routine care for the participants and collaborate with the GCKD study. A list of nephrologists currently collaborating with the GCKD study is available at http://www.gckd.org.
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Open Research Badges
Data Availability Statement
The code for modifying the RSF implementation is available at https://github.com/cbehning/ranger/tree/competing_risks_subdist. The code for replicating the simulation results is available at https://github.com/cbehning/rsf_competing_events
GCKD: Public posting of individual-level participant data is not covered by the informed patient consent form. As stated in the patient consent form and approved by the Ethics Committees, a dataset containing pseudonyms can be obtained by collaborating scientists upon approval of a scientific project proposal by the steering committee of the GCKD study: https://www.gckd.org.
This article has earned an Open Data badge for making publicly available the digitally-shareable data necessary to reproduce the reported results. The data is available in the Supporting Information section.
This article has earned an open data badge “Reproducible Research” for making publicly available the code necessary to reproduce the reported results. The results reported in this article were reproduced partially due to data confidentiality issues.