A Novel Eukaryote-Like CRISPR Activation Tool in Bacteria: Features and Capabilities
Abstract
CRISPR (clustered regularly interspaced short palindromic repeats) activation (CRISPRa) in bacteria is an attractive method for programmable gene activation. Recently, a eukaryote-like, σ54-dependent CRISPRa system has been reported. It exhibits high dynamic ranges and permits flexible target site selection. Here, an overview of the existing strategies of CRISPRa in bacteria is presented, and the characteristics and design principles of the CRISPRa system are introduced. Possible scenarios for applying the eukaryote-like CRISPRa system is discussed with corresponding suggestions for performance optimization and future functional expansion. The authors envision the new eukaryote-like CRISPRa system enabling novel designs in multiplexed gene regulation and promoting research in the σ54-dependent gene regulatory networks among a variety of biotechnology relevant or disease-associated bacterial species.
1 Introduction
CRISPR (clustered regularly interspaced short palindromic repeats) activation (CRISPRa) is a power and versatile technology for genetic engineering and biological research. The innate programmability from the CRISPR module offers unprecedented flexibility to turn on any target gene of interest. Its programmability was instrumental to CRISPR, which is an adaptive immunity in bacteria.[1-3] Prokaryotes have evolved mechanisms to protect themselves against exogenous DNA/RNA. They were performed by endonucleases that recognize the invasive species through RNA/DNA or RNA/RNA complementary pairing and then cleaving them.[4, 5] By exploiting the sequence-specific “identification friend or foe” system, scientists are able to guide the endonucleases to their desired DNA or RNA targets.[5-8]
CRISPR regulation relies on inactivated CRISPR endonucleases. The nuclease-deficient CRISPR DNA endonucleases, for instances dCas9 and ddCpf1 (dCas12), are effectively programmable DNA binding domains. When these domains are tethered to transactivation domains or subunits of RNA polymerase, they activate the promoters near the CRISPR target sites. This strategy has been widely utilized in both eukaryotes and prokaryotes,[9-17] particularly in the former, where the transcription activation mechanisms and the activators are well-studied.
While CRISPRa in eukaryotes enjoys much success and is continuously improved, the development of CRISPRa in prokaryotes had stagnated.[10, 15, 16] Bikard et al. reported the first bacterial CRISPRa device to employ an RNA polymerase ω-subunit fused dCas9 in a ω-deleted strain.[10] The second CRISPRa device has an E. coli activator SoxS, which was recruited to the dCas9 through the engineered single guide RNA (sgRNA) and an RNA-binding domain fused to the activator. SoxS interacts with the α-subunit of RNA polymerase. This enhances the activation efficacy and removes the prerequisite on host strain genetic backgrounds.[15] Recently, a dCas9-fused anti-sigma factor AsiA was employed for a CRISPRa system, which enriched the bacterial CRISPRa toolbox.[17]
The above introduced bacterial CRISPRa systems are designed for bacterial σ70-dependent genes, which perform most house-keeping functions.[18] CRISPRa systems for many other genes were still unavailable, for instance, many other biological functions that respond to environmental changes under the control of the σ54 factor, the only sigma factor apart from σ70 responsible for regulating various functions.[19] The σ54-dependent promoters have a distinct activation mechanism from its σ70 counterpart, with a unique promoter structure and its own set of conserved core sequences.[20] Their regulations work over long distances with a DNA looping structure, similar to that of the RNA polymerase II in eukaryotes. Hence, σ54 activation is also known as eukaryote-like gene activation in bacteria.[21-25] Much of the regulatory networks and biological functions of σ54-dependent genes remain elusive, a situation that hindered the standardization and application of σ54-dependent promoters in genetic engineering.
Recently, we developed a CRISPRa system for σ54-dependent genes. It supports high dynamic range regulation with low expression leakiness.[16] Thanks to the flexible DNA looping and the inherently long-distance regulation, CRISPRa target sites can be placed over a wider physical range. Furthermore, it enables direct activation of many σ54-dependent promoters, which otherwise have to be activated by environmental stimuli that would have global effects and complicate experimental control. These properties of our CRISPRa, together with our discoveries of CRISPRa design principles, will facilitate the research in σ54-dependent gene regulatory networks and the applications of σ54-dependent promoters in synthetic biology and industry.
We illustrated the application of the σ54-dependent CRISPRa by two examples. We built a two-layered cascaded activation and a positive feedback regulation. These circuits demonstrated the potential of our CRIPSRa to build complex gene regulatory networks. We also used a standardized σ54 promoter library to implement a high throughput method for screening the multi-gene expression profile. A multi-gRNA generator circuit was designed to optimize a multi-gene pathway by projecting various transcription profiles onto it. We envision that the success of our CRISPRa design and its application would open up exciting opportunities, and encourage other scientists to apply the eukaryote-like CRISPRa system for their scientific endeavors.
2 Existing CRISPR Activation Methods in Bacteria
To date, three canonical CRISPRa systems and one gene activation device using CRISPR-mediated DNA looping have been proven in bacteria. Here we briefly summarize their designs, properties, and their suitable applications (Table 1). The summary, therefore, is an abridged development history of bacterial CRISPRa. By compare and contrast, we would highlight the unique features and capabilities of our eukaryote-like CRISPRa system.
| Cas protein | Regulator | Activator attachment mode | Target promoters | Host cell background requirement |
|---|---|---|---|---|
| CRISPR/dCas9 | ω-subunit | Protein fusion/gRNA scaffold mediated | σ70 promoter | E. coli ΔrpoZ[10] |
| CRISPR/dCas9 | SoxS | gRNA scaffold mediated | σ70/38/32/24 promoter | None in E. coli[15] |
| CRISPR/dCas9 | AsiA (including mutant) | Protein fusion/gRNA scaffold mediated | σ70 promoter | None in E. coli, K. oxytoca, S. enterica (for protein fusion) & σ70 F563Y mutated E. coli (for gRNA scaffold mediated)[15, 17] |
| CRISPR/dCas9 |
PspFΔHTH NorR WtsA |
gRNA scaffold mediated | σ54 promoter | None in E. coli, K. oxytoca, E. coli ΔpspF (for enhanced function),[16] |
| CRISPR/dCas9 | σ factors | Protein fusion | σ70 promoter | None in M. xanthus[26] |
| CRISPR/dCas9 | TetD | gRNA scaffold mediated | σ70 promoter | None in E. coli[15] |
| CRISPR/dCas9 | - | DNA looping | σ70/54 promoter | None in E. coli[27] |
2.1 The ω-Subunit Based CRISPR Activation
Fundamentally, all CRISPRa systems capitalize on the natural modularity of genetic regulators or RNA polymerases, where separable domains are independently responsible for DNA binding, regulation, and recruitment of RNA polymerase or other transcription factors. These domains can be mixed and matched, and would trigger transcription activation as long as they are fused or assembled together to bring the RNA polymerase and accessory transcription factors to the vicinity of the promoter. This simple and yet robust characteristic is also the cornerstone of the two-hybrid system, which screens for molecular interactions in vivo: Candidate proteins/domains are fused to a DNA binding domain and an activation domain respectively. Any interaction between candidates would recruit the activation domain to the DNA binding domain and activate the promoter nearby, which converts interaction into observable gene expression.
The sufficiency of an activator-RNA polymerase interaction to initiate bacterial transcription was first reported in 1997. It was simultaneously the first reported bacterial two-hybrid (B2H) system that utilized the α-subunit of the RNA polymerase.[28, 29] This was followed by Dove and Hochschild in 1998, who showed that the ω-subunit of the RNA holoenzyme could also be used to recruit and stabilize the RNA polymerase to enhance transcription initiation.[30] This system, however, only works when the endogenous copy of the ω-subunit (rpoZ) has been knocked out.
The ω-subunit is an important component in RNA holoenzyme, which can respond to the alarmone ppGpp during stringent response and broadly regulate gene transcription. However, the ω-subunit (rpoZ) is not essential and its knockout does not significantly affect the E. coli host growth rate. Hence the function of this subunit was once vague and controversial for a long time.[31-33] The interaction between the ω-subunit and the other subunits of RNA polymerase is thought to contribute to its ability of transcription initiation.
Building on this information, Bikard et al. reported the first bacterial CRISPRa by fusing the ω-subunit to the dCas9.[10] The gRNA then brings the fused protein upstream of a constitutive promoter for activation (Figure 1a). Fusion can take place on the N or C terminal of dCas9. Since the ω-subunit is fused directly to dCas9, there is no additional design requirements to retrofit the gRNA structure. Activation at the optimal target site yielded a 23-fold dynamic range. The system works best on weak promoters—increasing the strength of the constitutive promoter decreases observable fold change in activation. Intuitively, strong constitutive promoters by definition already have high affinities with the RNA polymerase and additional recruitment through the ω-subunit would be insignificant.
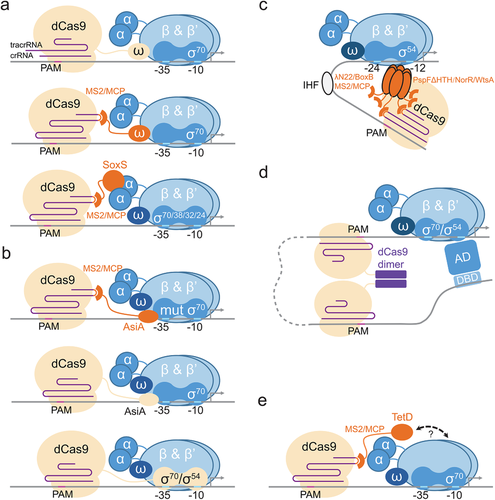
Bikard's CRISPRa system is highly sensitive to the distance between the CRISPR target site and the transcription start site (TSS), and they demonstrated a functional range of 20 bp. However, subsequent research using the phage-assisted continuous evolution (PACE) to evolve protospacer adjacent motif (PAM) compatibility-broad dxCas9 appeared to suggest a periodic variation of the activation efficiency along with the variation of the distance between TSS and the target site.[34] Such periodic variation of the activation efficiency that accompanies target site shift was also observed in both other bacterial CRISPRa systems, a feature that is likely a result of the molecular spatial orientation change owing to the periodic DNA helix structure. In another CRISPRa study, the RNA polymerase ω-subunit was fused to the MCP RNA adaptor, which can be recruited to target promoters using an engineered sgRNA scaffold with an MS2 RNA aptamer (Figure 1a). However, this design resulted in weak CRISPRa activation.[15]
2.2 The CRISPR Activation Based on the AraC/XylS Family Activators and Sigma Factors
The second CRISPRa system differs from the work by Bikard et al. in the activator, and the strategy in which the activator was linked to the dCas9/gRNA complex.[15] Dong et al. adopted the design from CRISPRa work in eukaryotic cells. The RNA aptamer MS2 is incorporated into the sgRNA scaffold, which recruits an RNA binding adaptor MCP domain fused to the activator (Figure 1a). This design has two advantages: 1) Recruitment of the activator through gRNA instead of direct fusion to dCas9 avoids unpredictable steric and functional interference to either of the two domains. 2) Different sgRNA on different target sites/promoters can now mediate CRISPRa or CRISPRi independently, depending on whether an activator is recruited to the sgRNA.[12]
To obtain a potent activation domain, the authors then screened several candidate bacteria activators, hijackers and RNA polymerase subunits, and identified the AraC/XylS family activators SoxS and TetD. SoxS yields the strongest activation efficiency and its CRISPRa system is functional in wild type E. coli, eliminating the need for gene knockouts in host strains. The two AraC/XylS family activators SoxS and TetD have different activation mechanisms.[35] Previous researches revealed that SoxS interacts with the α-subunit to activate transcription, while the interaction mechanism of TetD and RNA polymerase is still unclear[35-37] (Figure 1a,e). This suggested that this CRISPRa system may, in general, operate through the α-subunit. Yet surprisingly, using the α-subunit directly as the activator would not give significant CRISPRa activity, and the underlying reason is unclear. However, Peng et al. also reported that the dCas9 fused with an α-subunit has weak CRISPRa function in Myxococcus xanthus.[26] Therefore, α-subunit mediated CRISPRa might still be an option but further studies are needed.
In addition, Dong et al. discovered the T4 phage anti-sigma factor AsiA could act as an activator for CRISPRa when used in a strain expressing the F563Y σ70 mutant. AsiA normally inhibits transcription in E. coli by interacting with the σ70 factor.[38-40] This toxicity could be rescued by the σ70 mutant, which leaves the AsiA as merely a σ70-binding protein.[41] The CRISPR complex thus ultimately recruits the σ70 factor to boost transcription. Interestingly, in another recent design, the AsiA was fused directly to the dCas9, and toxicity was not observed, obviating the need of the F563Y σ70 mutant. When expressed as part of a fused protein, AsiA appeared to maintain its affinity with, but no longer inhibits the σ70 factor. The fact that direct fusions work for AsiA but not SoxS suggests that different activators require distinct engineering strategies depending on their mode of actions.[17]
In use of σ factors as activators, work by Peng et al. also reported functional but weak CRISPRa in Myxococcus xanthus could be obtained by fusing σ70, σ54, or the extracytoplasmic function (ECF) σ factor CarQ to dCas9[26] (Figure 1b).
Despite the innovation in design and the choice of activator, this CRISPRa system is similar to the one by Bikard et al., limited by the basal strength of target promoters. This might reflect a fundamental limitation in relying on RNA polymerase subunits to recruit and stabilize the RNA polymerases. Activation efficiency is also influenced by the sequences between the target site and the TSS.[42] Still, there might be an improvement in the distance between the target site and the TSS, since recent research indicates that a wider range can be tolerated. In addition, activation efficiencies appeared to alternate like sharp peaks as the distance between the TSS and target site increased.[42] This phenomenon might be due to the relative orientation of the activation complex as the binding site spirals along the DNA helix.
2.3 The Eukaryote-Like CRISPR Activation
The σ54-dependent promoters are a common but special group in bacteria. As the only promoter type that has a wide range of biological functions apart from the σ70-dependent promoters, σ54-dependent promoters generally control the genes that respond to environmental conditions.[19]
In contrast to σ70-dependent promoters, σ54-dependent promoters require activation for transcription initiation. The σ54 factor has special recognition sequence (–12 box and –24 box for σ54, while -10 box and -35 box for σ70) and a “blocking” mechanism to inhibit transcription initiation.[43, 44] When a σ54 activator complex binds to an upstream activating sequence (UAS), DNA looping occurs with the help of integration host factor (IHF) to facilitate its interaction with the σ54 factor.[45] This is followed by ATP hydrolysis, which provides the energy to switch off the “blocking” conformation of the σ54 factor to allow RNA polymerase initiating the transcription. Since the UAS is far away from the promoter, the regulation happens over long distances, and together with the iconic DNA looping structure, the activation mechanism shares similarity with that of the eukaryotic RNA polymerase II. Hence σ54 activation is known as the eukaryote-like activation in bacteria.[21-25, 46]
The above properties were taken into consideration in the design of our CRISPRa based on σ54-dependent promoters, and we also drew insights from a σ54-based B2H system.[47] Since the σ54 activator functions as a hexamer, it could not be fused to the dCas9 protein directly and has to be recruited through an engineered gRNA with at least two RNA aptamers.[16] The original DNA binding domains in the activator hexamer were deleted and replaced by RNA binding adaptors (Figure 1c). This allows the engineered activators to be recruited to the CRISPR complex and at the same time precludes their activation of native target genes on the genome. By screening several σ54 activators from different bacterial species, three σ54 activators (PspF, NorR, WtsA) were identified that could be engineered to be activators specific for CRISPRa. During this process, we noted that our candidate σ54 activators are highly diverse and showed little modularity in domain functional structures. We concluded that the activator for the phage shock protein operon (pspABCDE) PspF is the most potent activator.[16, 48-50]
The novel use of σ54-dependent promoters for CRISPRa overcame some issues faced by other bacterial CRISPRa system. This system not only works in the σ54 activator pspF gene knockout and wild type E. coli, but also in a non-model bacterium Klebsiella oxytoca. There is no restriction on the strength of the target promoter, because all the σ54-dependent promoters are blocked and are effectively at “OFF” states unless activated. With the long-distance regulation mechanism, the target site can be placed much farther away from the TSS. Our thorough characterization between target site-TSS distance and activation efficacy revealed a waveform alternation, demonstrating for the first time a clear spatial relationship at work in CRISPRa.
2.4 CRISPR Activation by DNA Structure Remodeling
The three methods above rely on tethering an activation domain to the CRISPR complex. The fourth method deviated from this norm by omitting the activator domain, and achieve activation solely by DNA looping. The dCas9 was engineered to form dimers and two monomers were targeted to a distantly separated activator binding site and the promoter.[27] Dimerization of the bound dCas9 thus brings the two elements into proximity and reconstitutes a local activated promoter (Figure 1d). Theoretically, this method could activate both σ70 and σ54 promoters possessing available activation mechanisms, and it is not limited by the genetic background of the host. A recent study indicates that the E. coli chromosome is well-mixed and uncompartmentalized, further supporting the application potential of this strategy.[51] However, the dynamic range of this strategy is lower than other reported methods, and would require much optimization if a further application is desired.
3 Features of Eukaryote-Like CRISPR Activation
The eukaryote-like CRISPRa system has many unique features because of its σ54-dependent activation mechanism, some of which have important value for application in synthetic biology and industrial production. The following section reviews the features and the mechanisms behind.
3.1 Low Background Expression and High Dynamic Range
The key principle underlying σ54-dependent CRISPRa is quite different from others. In σ70-dependent CRISPRa, the target σ70 promoters have their constitutive transcription initiation efficiencies. Activation by the CRISPRa system enhances the transcription initiation rates by stabilizing and recruiting RNA polymerase onto promoters. Whereas for σ54 promoters, activation is the release of a tightly locked promoter activity. This minimizes the waste of cellular resources when gene expression is unnecessary.
The transcription output of an activated σ54-dependent promoter can be very strong. This property has been harnessed in synthetic biology for the construction of digital-like genetic logic gates and analog signal amplifiers, which optimized biosensors performances with programmed selectivity and sensitivity.[52-56] Our eukaryote-like CRISPRa also benefited from this feature and thus has high dynamic ranges in inducible regulation, which further enabled applications that were otherwise impossible.[16]
In our CRISPRa system, in the absence of dCas9, sgRNA and the activator, background output is almost undetectable. It is only observed when the activator is constitutively expressed and causes non-specific activation. This may be explained by random molecular interactions between the RNA polymerase and the engineered activator. Interestingly, some rare σ54-dependent activators lack the DNA binding domains and hence rely solely on their own high expression levels to activate target promoters.[57]
3.2 A Wider Region for Target Site Selection
In σ54-dependent promoter activation, there is little limitation on the distance between the UAS and the core promoter. In one case, the UAS could be 1 kb away from the TSS.[58] The interaction between the activator and the RNA polymerase is made possible by the eukaryote-like DNA looping structure. In our CRISPRa system, this flexibility in molecular structure confers flexibility in choosing a target site. Within the 40 bp region that we tested, all target sites were functional. All CRISPR-derived systems require the presence of PAM. PAM, as its name depicts, is a short sequence right next to the target DNA region and is the first recognized element in CRISPR surveillance.[3, 59] Our CRISPRa system increases the chance of finding one because a wider sequence space could be used. Moreover, the presence of multiple operable target sites on the same promoter implies that programmable, multi-input logic gates can be realized by our CRISPRa system. This property supports higher degrees of freedom for PAM site choosing and programmable multi-input CRISPRa-enabled logic gate design.
4 Design Principles Learned from the Eukaryote-Like CRISPRa
Below we summarize the design principles learned during the construction and optimization of our CRISPRa system. This should aid the future use of our system by others.
4.1 PAM Site Position is Vital for Artificial Target Promoter Design
Our CRISPRa system shows an alternating change in the activation efficiency as the target site slides along the DNA strand, at a rate of one cycle every 10 bp (Figure 2a). This indicates a spatial relationship between the activator and the RNA polymerase along the DNA double helix.[16] The CRISPR-activator complex likely spirals along the DNA helix and result in alternating displacement between the recruited RNA polymerase and the promoter, and hence the efficiency (Figure 2a). This finding has important implications in predicting the output of our CRISPRa system. Recently, similar observations were reported in other CRISPRa systems, but the pattern was not identical to ours, which may be explained by differences in mechanisms.[42]
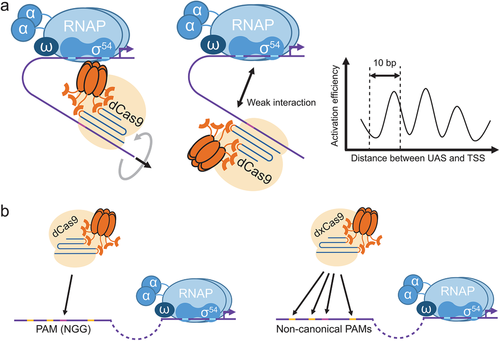
In our study, we opted for distances of −127 and −107 bp and obtained an orthogonal library of synthetic CRISPR activatable promoters that performed well. Future users are recommended to observe these distances for optimal CRISPRa effects on pspA promoter.
4.2 sgRNA Scaffold Design Affects the Function of Eukaryote-Like CRISPRa
For CRISPRa devices utilizing the strategy of the gRNA-recruited activation domain, the structure of the engineered sgRNA could affect the function of the CRISPRa system. The design of our eukaryote-like CRISPRa shows both quantity and location preference of RNA aptamers in the sgRNA scaffold. Notably, some specific stem-loop structures could not be engineered and combined as RNA aptamers to achieve functional CRISPR activation. Due to the underlying diversity of different CRISPRa systems, it is unknown whether this phenomenon is common for all bacterial CRISPRa devices. In summary, it suggests that the gRNA scaffold structure could be an important factor to affect the function of CRISPRa.
4.3 dCas9 Mutants Enable PAM Preference Expansion
Locating a PAM is a prerequisite for using the CRISPRa system. This could pose a challenge for endogenous gene activation, since a functional PAM (NGG for SpdCas9) may not be present on the wild type promoter sequence. The problem could be solved by employing some dCas9 variants that target sequences with non-canonical PAM. In 2018, Hu et al. generated a number of dxCas9 that recognize different non-canonical PAM.[34] We utilized the variant dxCas9 3.7 to activate the gene for phage shock protein pspA on the E. coli genome[16] (Figure 2b). We noticed that a similar strategy was subsequently employed on another σ70-dependent CRISPRa system.[42]
dxCas9 is not the only option. Nishimasu et al. rationally designed the mutant Cas9 SpCas9-NG, which efficiently cleaves DNA with non-canonical PAM.[60] A nuclease-deficient version of Cas9-NG would enable non-canonical PAM recognition in our CRISPRa system. In addition, other natural CRISPR/Cas9 proteins have their PAM preferences as well, which could be utilized to expand the PAM preferences of CRISPRa systems.[61-63]
4.4 Digital Response by Feedback Regulation
In synthetic biology, the digital response is a property that is helpful in signal amplification and cascade control,[64-66] and is frequently achieved by the introduction of a positive feedback loop that steepens the response curve.[67-71] Construction of such loops using CRISPRa in bacteria has long been a challenge due to insufficient dynamic range and limitation of high basal expression. Since our CRISPRa overcame such issues, we were able to implement a positive feedback loop to achieve a digital response.[16] We foresee this strategy to be of interest in future research.
5 What does Eukaryote-Like CRISPRa Allow Us to Do?
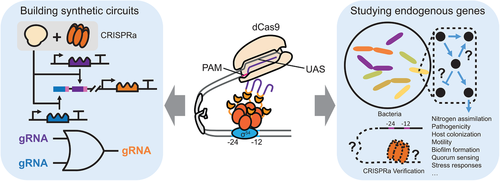
CRISPRa technologies are well-posed to address two issues: 1) Regulation of endogenous genes and 2) Construction of synthetic gene circuits (Figure 3). Below we discuss the areas in which our eukaryote-like CRISPRa could contribute, the advantages it offers, and the potential limitations it may face.
5.1 Investigation of the “Dark Matter”—σ54-Dependent Gene Regulatory Networks
σ54-dependent promoters generally control genes that respond to changes in environmental conditions, and apart from the σ70-dependent promoters, they are the only class of promoters that is not associated with a specific programmed response but controls over various biological functions.[19] This is supported by the genome-wide DNA binding study that had identified 135 binding sites for σ54 from the E. coli genome,[72] and an analysis of 186 σ54 promoters and putative σ54 promoters from 47 species.[73] For instances, σ54 promoters are involved in nitrogen assimilation and fixation,[74-76] pathogenicity,[77-79] host colonization,[80] motility,[78, 80, 81] biofilm formation,[80] quorum sensing,[82, 83] stress responses,[76] and environmental bioremediation[84] (Figure 3).
Basic research of σ54-dependent promoters is crucial for disease control. The Lyme disease affects about 300 000 people every year in the USA, and it has been shown that σ54 activation in Borrelia burgdorferi plays a pivotal role in the infection and transmission process.[78, 85] Virulence in many other pathogenic bacteria such as Helicobacter pylori, Burkholderia cenocepacia, Erwinia stewartii (a plant pathogen), Vibrio fischeri (an aquatic pathogen) is also controlled by gene regulatory networks that are σ54-dependent.[77, 80, 86, 87]
Many putative σ54-dependent promoters were identified from systems biology and bioinformatics studies, but most remained unverified due to the complex regulatory background and the lack of molecular tools for precise genetic perturbation. There are two major reasons behind the difficulty to develop those much needed tools.
First, σ54-dependent promoters have very diverse structures. In most cases, the only instantly identifiable elements are the core regions, the –12 box and –24 box, which are highly conserved. Regions for DNA looping often have different lengths, and the UAS for different σ54 activators share little homology with one another.[88]
Second, some σ54-dependent genes require uncommon laboratory conditions to activate. The provision of such conditions may require substantial investment and commitment, and thus hinders the research of those genes. For example, the gene cluster for nitrogen fixation in K. oxytoca is σ54-dependent, and would only turn on under anaerobic conditions.[89, 90]
These limitations can be overcome using the σ54-dependent CRISPRa system. This system can orthogonally activate σ54-dependent genes, and therefore assist in the research of their functions. We demonstrated the proof of concept on two key promoters in the nitrogen fixation pathway in K. oxytoca, and successfully activate them under an aerobic condition.[16] Furthermore, our CRISPRa system can also verify putative σ54-dependent promoters by targeting the CRISPRa complex to the putative UAS.
5.2 Multi-Gene Expression Profile Control
The programmability of CRISPR derived system facilitates multiplexed gene regulation. Our CRISPRa system is uniquely suitable for this task given its high dynamic range, stability, and durability. One area of our applications is to accelerate the construction and screening of metabolic pathway expression profiles. Such processes are often laborious and time-consuming. Typically, the identification of an optimal expression profile for a new metabolic pathway requires the construction of a new profile library, and if the library confers cytotoxicity and burden, it will have to be rebuilt. To address this issue, we designed a library with multiple gRNA constitutively expressed, and in different profiles, the same set of gRNA with different strengths of expression are combined combinatorically (Figure 4a). When projected through our CRISPRa system, the different production rates of the gRNA set would translate into different expression levels of the genes in the target pathway that have corresponding orthogonal σ54-dependent promoters (Figure 4b,c). The intensity of projection is tuneable and the same gRNA generator library could be reused in the optimization of a new metabolic pathway.

5.3 Design of Cellular Computing Devices
CRISPRa-mediated multi-layered cellular computing devices have achieved much success in eukaryotes,[66, 91, 92] but so far no such examples have been reported in bacteria. This is likely due to the low dynamic ranges of bacterial CRISPRa in the past, which failed to support cascaded regulation. In addition, no more than one UAS can be functional in one promoter, so signal integration and hence multi-input regulation is difficult to achieve. These two limitations do not concern the eukaryote-like CRISPRa system, and we reported a CRISPRa-based cascaded circuit that paves the way for high-level cellular computing.[16]
6 Potential Future Directions for the Eukaryote-Like CRISPRa System
Our observations on our CRISPRa system led us to propose the following ideas for further experimentation and functional expansion of the system.
6.1 Activator Optimization by Directed Evolution
Directed evolution has been used to generate a dxCas9 variant and to optimize a bacterial CRISPRa system.[17, 34] This strategy is universal and would be valuable in improving our eukaryote-like CRISPRa system. The activator and the sgRNA scaffold could be evolved to yield more potent variants. Selection pressure can be set up easily by coupling activation efficiencies to cell survivability through the expression of a selection marker.
6.2 The Necessity of DNA Looping and Availability of Other CRISPR/Cas Proteins
In σ54-dependent promoters, the DNA segment between the promoter core region and the enhancer forms a loop structure when the activator interacts with the RNA polymerase. Often, loop formation is mediated by the IHF DNA bending protein. The question is whether the loop is the prerequisite or the effect of the activator-RNA polymerase interaction. If it is the former, then there exists a possibility to satisfy that spatial requirement through an artificially designed flexible structure.
In a previous study, IHF-independent activation of σ54-dependent promoters could be achieved by substituting the DNA loop fragment with sequences that are inherently more flexible.[45] This implies that the loop serves to bend the DNA such that the bound activator and RNA polymerase can come into proximity, and if the straining DNA is out of the picture, the affinity between the activator and the RNA polymerase suffices to drive activator-RNA polymerase interaction. Hence, we hypothesize that a long flexible RNA or peptide linker between the activator and the CRISPR/Cas complex may provide sufficient spatial degrees of freedom for the activator-RNA polymerase interaction, and hence bypass the DNA looping requirement. This flexibility could mean that the activator anchors on gRNA scaffold can be relocated to other parts of the gRNA and would no longer be limited to the gRNA structure from Streptococcus pyogenes. If that is possible, the dCas9:gRNA complex in the CRISPRa system might be further substituted by other CRISPR/Cas systems, for instance, dCas12.
6.3 Potential Eukaryote-Like CRISPRa on σ70-Dependent Promoters
We envision that the eukaryote-like CRISPRa system may be modified to activate σ70-dependent promoters. In a special case, the NtrC-family regulator PhhR, a homolog of the σ54 activators, activates a σ70-dependent promoter by long distance regulation.[93, 94] PhhR has been suggested to interact with the α-subunit through an IHF-dependent DNA looping structure. This model may provide a basis to engineer a eukaryote-like σ70-dependent CRISPRa system.
7 Conclusions and Prospects
CRISPR activation in bacteria has seen much development in the last 7 years. There are two main strategies in creating a bacterial CRISPRa system. One is designing an engineered CRISPR/dCas9 complex that interacts with and stabilizes the RNA polymerase holoenzyme on weak σ70 promoters. The other is the eukaryote-like CRISPRa, which recruits a σ54 activator to a σ54-dependent promoter through the CRISPRa complex. These two strategies concern two different types of promoters and therefore are difficult to be cross-applied to promoters of the other type.
The eukaryote-like CRISPRa system is applicable to activating endogenous genes and constructing synthetic circuits. The optimization strategy for each application will differ according to their specific goals. For endogenous gene activation, being flexible in choosing the location of a target site and satisfying PAM requirements are paramount. Using dCas9 variants that target non-canonical PAM would be helpful. Whereas in the construction of synthetic circuits, dynamic range is the key to performance. Our eukaryote-like CRISPRa is well-suited given its high dynamic range and low background output. We further worked out design principles that serve as guidelines in the optimization of dynamic ranges. We believe the future in CRISPR-based circuits is “CRISPR regulation Supremacy,” where CRISPR regulation can build complex genetic circuits that are unachievable using non-programmable gene regulatory devices.
Programmable gene activation is a promising, yet highly competitive and challenging research field. The eukaryote-like CRISPRa system fills the long-neglected void of programmable σ54-dependent promoter activation, and will expedite the research σ54-dependent gene regulatory networks. On the other hand, our system is a novel tool for synthetic circuit design and construction in synthetic biology. Given a strong interest, we believe existing CRISPRa technologies will be continuously improved, and more CRISPRa devices and applications will emerge in the near future.
Acknowledgements
The authors thank Trevor Y. H. Ho for his help in preparing the manuscript and valuable suggestions. This work was supported by UKRI Future Leaders Fellowship [MR/S018875/1], UK BBSRC grant [BB/N007212/1], Leverhulme Trust grant [RPG-2015-445], and Wellcome Trust Seed Award in Science [202078/Z/16/Z].
Conflict of Interest
The authors declare no conflict of interest.




