Tuning Encodable Tetrazine Chemistry for Site-Specific Protein Bioorthogonal Ligations
Graphical Abstract
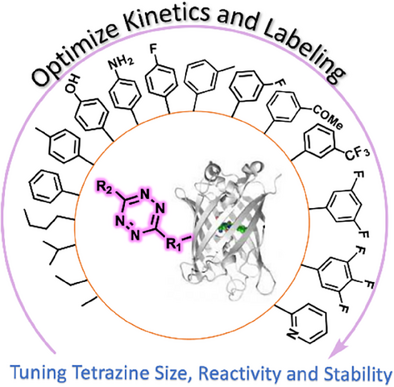
We expand the bioorthogonal chemistry toolkit by synthesizing and characterizing 29 tetrazine ncAAs and evaluating their genetic encoding into proteins. Site-specific incorporation was achieved, and tools to assess efficiency, fidelity, stability, and reactivity were applied. This roadmap enabled the design of tunable tetrazines and the fastest quantitative labeling at 10⁶ M⁻¹s⁻¹ without compromising stability.
Abstract
Using genetic code expansion (GCE) to encode bioorthogonal chemistry has emerged as a promising method for protein labeling, both in vitro and within cells. Here, we demonstrate that tetrazine (Tet) amino acids incorporated into proteins are highly tunable and have extraordinary potential for fast and quantitative bioorthogonal ligations. We describe the synthesis and characterization of reaction rates for 29 Tet amino acids (20 of which are new) and compare their encoding ability into proteins using evolved tRNA/RS pairs. For these systems, we characterized on-protein Tet stability, reaction rates, and ligation extents as the utility of a bioorthogonal labeling group depends on its stability and reactivity when encoded into proteins. By integrating data on encoding efficiency, selectivity, on-protein stability, and in-cell labeling for Tet tRNA/RS pairs, we developed the smallest, fastest, and most stable Tet to date. This was achieved by introducing fluorine substituents to Tet4, resulting in reaction rates at the 10⁶ M⁻¹s⁻¹ level while minimizing degradation. This study expands the toolbox of bioorthogonal reagents for Tet-sTCO-based, site-specific protein labeling and demonstrates that the Tet is a uniquely tunable, highly reactive, and encodable bioorthogonal functional group. These findings provide a foundation to further explore Tet encoding and reactivity.
Open Research
Data Availability Statement
The data that support the findings of this study are available in the Supporting Information of this article.