Histone Deacetylase 3-Directed PROTACs Have Anti-inflammatory Potential by Blocking Polarization of M0-like into M1-like Macrophages
Minor changes have been made to this manuscript since its publication in Early View.
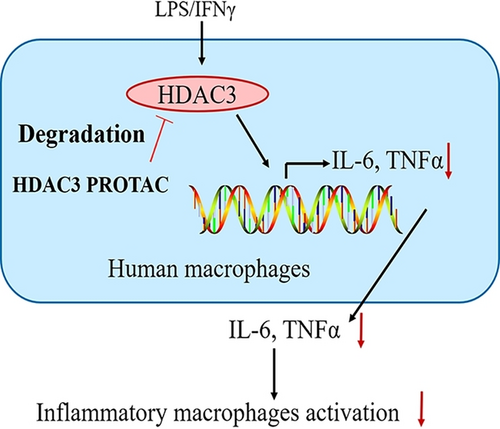
Graphical Abstract
We employed the proteolysis targeting chimera (PROTAC) technology to target the non-enzymatic histone deacetylase 3 (HDAC3) function. A newly developed potent and selective HDAC3 PROTAC provided an anti-inflammatory cytokine expression profile in cultured cells and inhibited the pro-inflammatory M1 phenotype in primary macrophages.
Abstract
Macrophage polarization plays a crucial role in inflammatory processes. The histone deacetylase 3 (HDAC3) has a deacetylase-independent function that can activate pro-inflammatory gene expression in lipopolysaccharide-stimulated M1-like macrophages and cannot be blocked by traditional small-molecule HDAC3 inhibitors. Here we employed the proteolysis targeting chimera (PROTAC) technology to target the deacetylase-independent function of HDAC3. We developed a potent and selective HDAC3-directed PROTAC, P7, which induces nearly complete HDAC3 degradation at low micromolar concentrations in both THP-1 cells and human primary macrophages. P7 increases the anti-inflammatory cytokine secretion in THP-1-derived M1-like macrophages. Importantly, P7 decreases the secretion of pro-inflammatory cytokines in M1-like macrophages derived from human primary macrophages. This can be explained by the observed inhibition of macrophage polarization from M0-like into M1-like macrophage. In conclusion, we demonstrate that the HDAC3-directed PROTAC P7 has anti-inflammatory activity and blocks macrophage polarization, demonstrating that this molecular mechanism can be targeted with small molecule therapeutics.
Introduction
Macrophages play a crucial role in innate and adaptive immune responses to pathogens and are also critical mediators of inflammatory processes. Macrophages can exhibit both pro- and anti-inflammatory functions.1-4 Based on the phenotype, macrophages can be divided into two distinct polarized states: classically activated macrophages (M1) and alternatively activated macrophages (M2).1, 2, 5, 6 The classically activated macrophages (M1) are induced by type 1 helper (Th1) cytokines such as interferon-gamma (IFNγ), and Toll-like receptor (TLR) ligands such as lipopolysaccharide (LPS). Although M1 macrophages are beneficial for host protection, unrestrained inflammatory macrophage activity aggravates and sustains chronic inflammatory diseases such as atherosclerosis, multiple sclerosis and rheumatoid arthritis.1, 2, 4 Alternatively activated macrophages (M2) are induced by various non-inflammatory factors, including the type 2 helper (Th2) cytokines interleukin-4 (IL-4), interleukin-13 (IL-13), interleukin-10 (IL-10), transforming growth factor-beta (TGF-β), glucocorticoids, and immune complexes. M2 macrophages have anti-inflammatory immunoregulatory functions that are beneficial during chronic inflammatory diseases.1-3, 6 Therefore, prevention of M1 macrophage polarization using small-molecule modulators may be a promising strategy for the treatment of inflammatory diseases.
Enzymes involved in the regulation of post-translational acetylation of histone lysine residues have been recognized as potential drug targets in cancers, neurological disorders, inflammatory diseases, and infection.7-9 Histone lysine acetylation is regulated by histone deacetylases (HDACs) as erasers of acetylations and by histone acetyltransferases (HATs) as writers of acetylations on histone lysine residues.8, 9 Selectivity can be gained by targeting isoenzymes of the HDAC family, which consists of eleven zinc-dependent HDACs grouped into three classes: class I (HDAC1,2,3, and 8), class II (HDAC4,5,6,7,9 and 10), and class IV (HDAC11) and seven NAD+-dependent HDACs grouped in class III (SIRT1-7).7-9 Importantly, biochemical studies have demonstrated that HDACs also exert non-enzymatic functions that cannot be targeted by inhibition of their deacetylase activity.10, 11 This calls for the development of new modalities to interfere with non-enzymatic HDAC functions.
Although HDAC inhibitors were initially developed for cancer therapy, several reports have demonstrated their potential for anti-inflammatory therapy.6, 12-14 Despite their anti-inflammatory potential, several studies demonstrated a pro-inflammatory effect upon HDAC inhibition.15-18 Recent studies provided more insight into the role of HDAC3 in inflammation. For example, Chen et al. found that HDAC3-deficient macrophages were unable to activate almost half of the inflammatory gene expression repertoire when stimulated with LPS.19 Our team found that siRNA-mediated HDAC3 knockdown led to upregulation of the expression of the anti-inflammatory cytokine IL-10 in LPS/IFNγ-treated RAW 264.7 macrophages.20 However, the HDAC3-selective inhibitor, RGFP966, did not show a significant impact on gene expression.20 Additionally, Mullican et al. reported that macrophages lacking HDAC3 displayed a polarization phenotype similar to IL-4-induced alternative activation and are hyperresponsive to IL-4 stimulation.21 In a more recent effort, Nguyen et al. have discovered that HDAC3 is a dichotomous transcriptional activator and repressor, with a non-canonical deacetylase-independent function. HDAC3 activates LPS-stimulated pro-inflammatory gene expression in a deacetylase-independent manner.10 In addition, they also discovered that the deacetylase-dependent and deacetylase-independent functions of HDAC3 have opposing roles in the toxicity of LPS and the toxicity can be magnified in the absence of its deacetylase activity.10 As a consequence, it becomes apparent that blocking the deacetylase-independent function of HDAC3 is also an important strategy for the development of therapeutics for inflammatory diseases.
Proteolysis targeting chimeras (PROTACs) have emerged as a new drug modality to induce degradation of the protein of interest (POI). PROTACs are heterobifunctional molecules consisting of a POI-binding ligand, an E3 ligase-recruiting ligand, and a linker.22-24 This heterobifunctional character enables simultaneous binding to the POI and the E3 ligase to form a ternary complex, thus leading to targeted protein degradation via the ubiquitin-proteasome system (UPS).22, 23, 25, 26 The event-driven PROTACs enable targeting of non-enzymatic protein functions that are undruggable for conventional occupancy-driven therapeutics.27-29 Moreover, the PROTACs act catalytically, which enables PROTACs to produce more sustained pharmacodynamic effects in a sub-stoichiometric manner.30, 31 Thus, the PROTAC strategy has the potential to target non-catalytic HDAC functions.
In a previous attempt, our group reported an HDAC3-selective PROTAC, denoted HD-TAC7, that included a 2-aminobenzamide core that was tethered to the cereblon (CRBN) E3 ligase ligand pomalidomide.32 However, HD-TAC7 mediated HDAC3 downregulation failed to enhance gene expression of the anti-inflammatory cytokine IL-10, in contrast to siRNA-mediated HDAC3 downregulation, which clearly enhanced IL-10 expression.20, 32 Given that nuclear factor-kappa B (NF-κB) p65 is downregulated by pomalidomide and that this transcription factor is involved in IL-10 expression, we reasoned that the failure of HD-TAC7 to increase IL-10 gene transcription could be ascribed to the CRBN ligand pomalidomide.32 This provides a rationale to replace the CRBN ligand with the von Hippel-Lindau (VHL) E3 ligase ligand, which is another widely used E3 ligase in PROTAC design.33, 34
In the current work, we developed two novel series of VHL-based HDAC3 PROTACs by linking HDAC3 deacetylase inhibitors to the VHL E3 ligase ligand. Based on the structure–activity relationships the potency and selectivity were optimized to provide the HDAC3-directed PROTAC P7. Its effect on expression of pro-inflammatory and anti-inflammatory genes was analyzed in both THP-1-derived macrophages and human primary macrophages. Subsequently, enzyme-linked immunosorbent assay (ELISA) and flow cytometry assays were conducted to evaluate the effects of PROTAC P7 on pro- and anti-inflammatory cytokine production and macrophage polarization based on human primary macrophages, respectively. Altogether, development of this PROTAC enables the inhibition of a pro-inflammatory cytokine expression profile and macrophage polarization, thus indicating the potential of HDAC3-directed PROTACs to explore HDAC3 pharmacology as a potential drug target in inflammation.
Results and Discussion
PROTAC design
The development of the HDAC3-directed PROTAC started with compound 1 (BRD3308, Figure 1) as a ligand with HDAC3-selectivity over HDAC1 and HDAC2.35 The known pharmacophoric model of HDAC inhibitors36, 37 and a docking study (Figure S1A) justify the presumption that the 2-aminobenzamide group is deeply embedded in the active site of HDAC3, where it functions as zinc-binding group (ZBG) and forms multiple hydrogen bond interactions with key amino acid residues. The acetyl group protrudes out of the pocket and is solvent-exposed, indicating that the attachment of linkers might be tolerated in this position. Subsequently, we proposed a binding mode between compound 2 (VHL-021, Figure 1)38 with VHL E3 ligase (Figure S1B), in which 2 is involved in hydrogen bond interactions with residue Try-98 and His-110 in the active pocket. Also here the acetyl group projects out of the pocket, thus indicating that this position is suitable for linker attachment. The P1-P5 series of HDAC3-directed PROTACs were designed accordingly by tethering HDAC3 inhibitor 1 to VHL E3 ligase ligand 2 via aliphatic linkers of various length (Figure 1). The synthetic Scheme is shown in Scheme S1.
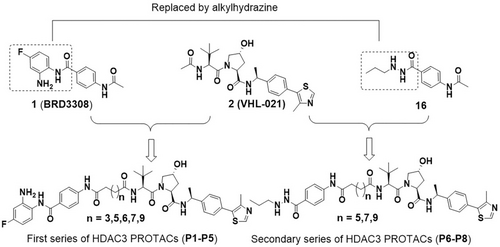
Conceptual design of VHL-based HDAC3 PROTACs.
PROTACs P1-P5 provide limited HDAC3 degradation
Binding of PROTACs P1-P5 to HDAC1, 2 and 3 was verified and their ability to induce HDAC3 degradation was evaluated. The IC50 values for HDAC3 inhibition ranged between 76 nM to 2.7 μM (Table 1 and S1). Interestingly, the linker length showed a significant impact on HDAC3 inhibition. Shortening the linker length increased the inhibitory activities against HDAC3. However, also the HDAC3 inhibitory selectivity over HDAC1 and HDAC2 seems to be reduced in comparison to HDAC inhibitor 1. Overall, this indicates that P2-P5 retain their HDAC3 binding potential.
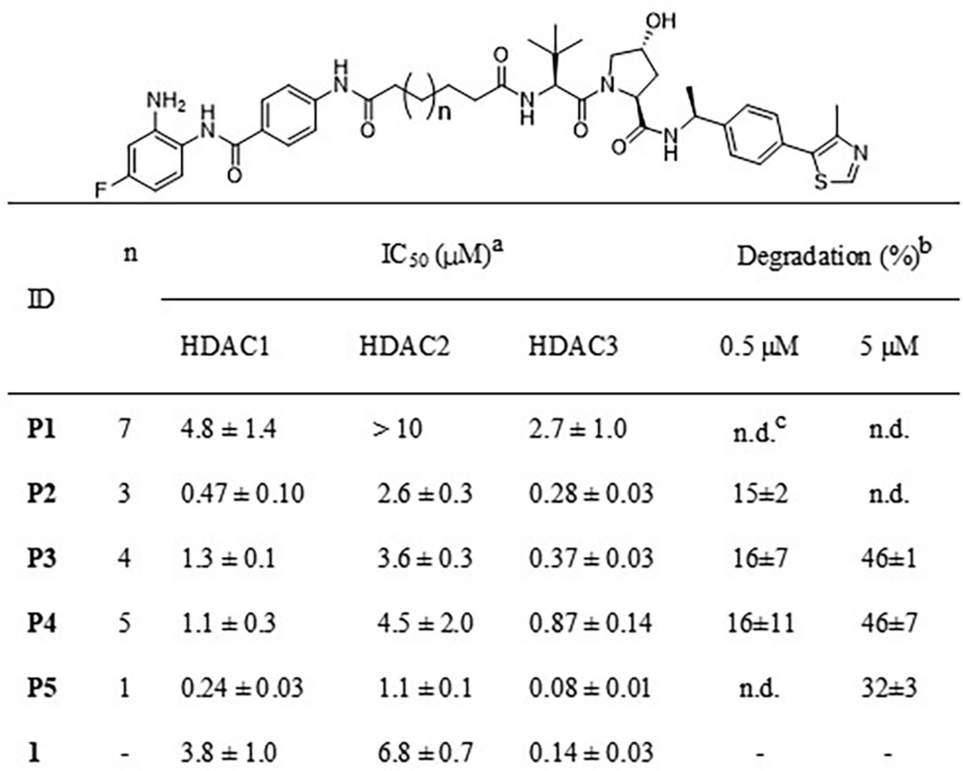
- [a]Inhibitory potency of compounds against HDAC1, 2 and 3 deacetylase activity, values are shown as mean ± SD, n=2 independent experiments. [b]HDAC3 degradation percentage is represented as mean ± SD, n=3 independent experiments. [c]No degradation.
The ability of PROTACs P1-P5 to induce HDAC3 degradation was investigated in the THP-1 cell line, which is a human leukemia monocytic cell line and has become a common model to estimate the modulation of monocyte and macrophage activities.39 THP-1 cells were treated with two different PROTACs concentrations (5 μM and 0.5 μM) for 24 h to anticipate on the hook effect that is commonly observed in PROTAC treatment.40, 41 The HDAC3 levels were determined via western blot analysis (Table 1 and Figure 2). At a concentration of 5 μM, PROTACs P3-P5 decreased HDAC3 levels by 32 % - 46 % compared to vehicle control, while PROTACs P1-P2 did not trigger HDAC3 degradation (Figure 2). However, upon treatment with 0.5 μM P2, P3 or P4 the HDAC3 levels were reduced by just 15 % - 16 %, whereas no degradation was observed for P1 and P5 (Figure 2). Overall, the ability to induce HDAC3 degradation in THP-1 cells appeared to be limited for this series of PROTACs.
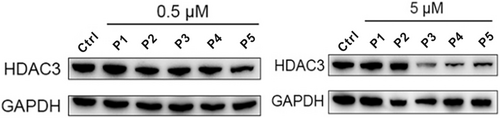
THP-1 cells were treated with 0.5 μM or 5 μM of PROTACs P1-P5 for 24 h. HDAC3 level was detected using western blot. GAPDH was used as a loading control. The corresponding uncropped images for Figure 2 are shown on page 25 in the Supporting Information.
Development of a Novel Series of HDAC3 PROTACs with Improved Potency
In order to improve the HDAC3-degrading potency, we searched for a replacement of the HDAC3 binding ligand. Recently, the acylhydrazine core has been identified as a zinc-binding group to replace the 2-aminobenzamide or hydroxamic acid. This provides enhanced potency for class I HDACs, especially for HDAC3.42-45 In addition, the hydrazide-based HDAC inhibitors show unique fast-on/slow-off HDAC-binding kinetics.42 Therefore, the 2-aminobenzamide moiety of compound 1 was replaced with acylhydrazine to provide compound 16 (Figure 1), which is expected to be a more potent HDAC3 inhibitor. The chemical synthesis of compound 16 is shown in Scheme S2 and the ability to inhibit the HDAC1, 2 and 3 was measured (Table 2 and Table S2). In line with the literature,43-45 compound 16 has 4-fold increased inhibitory activity against HDAC3 (IC50=30 nM) compared to compound 1, despite its decreased selectivity over HDAC1/2. This indicates that compound 16 can be used as HDAC3 ligand for PROTAC design.
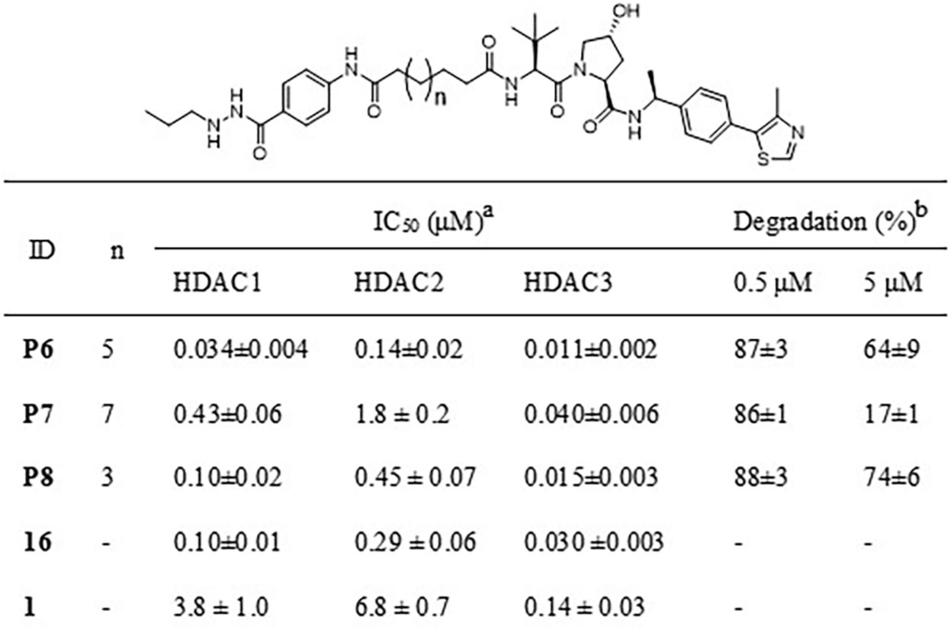
- [a]Inhibitory potency of compounds against HDAC1, 2 and 3 deacetylase activity, values are shown as mean ± SD, n=2–3 independent experiments. [b]HDAC3 degradation percentage is represented as mean ± SD, n=2 independent experiments.
The second series of HDAC3-directed PROTACs was assembled using 16 as an HDAC3 ligand. Compound 16 was linked to compound 2 via aliphatic linkers of various length to provide PROTACs P6, P7 and P8 (Scheme S3). These PROTACs provided IC50 values for HDAC3 inhibition between 11 nM to 40 nM (Table 2), which is in the same range as the IC50 of 16. Among these three PROTACs P7 has the best selectivity towards HDAC3 in comparison to HDAC1 and 2 with a difference in IC50 of a factor 10 or more (Table 2 and Table S2). Subsequently, the ability of PROTACs P6-P8 to induce HDAC3 degradation in THP-1 cells was determined. All PROTACs of this series efficiently triggered HDAC3 degradation and decreased HDAC3 levels by over 85 % at 0.5 μM (Table 2 and Figure 3A). The HDAC3 degradation-inducing ability at 5 μM decreased most clearly for P7, most likely due to the hook effect. Importantly, no obvious degradation of HDAC1 and HDAC2 was observed upon treatment with 0.5 μM or 5 μM of the PROTACs P6-P8, thus indicating selectivity among these HDACs (Figure 3A). Furthermore, the PROTACs P6-P8 also induced HDAC3 degradation in HeLa cells at 0.5 μM without obvious impact on the levels of HDAC1 and 2 (Figure 3B). Taken together, this series of PROTACs P6-P8 demonstrated excellent inhibitory potency against HDAC3, and effectively and selectively triggered HDAC3 degradation in both THP-1 cells and HeLa cells.
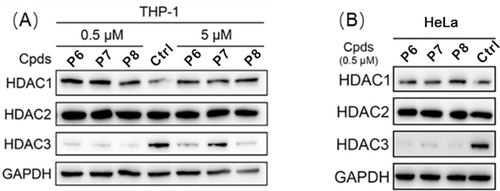
(A) THP-1 cells were treated with 0.5 μM or 5 μM of PROTACs P6-P8 for 24 h. The levels of HDAC1, 2 and 3 were detected using western blot. GAPDH was used as a loading control. (B) HeLa cells were treated with 0.5 μM of PROTACs P6-P8 for 24 h. The levels of HDAC1, 2, and 3 were detected using western blot. GAPDH was used as a loading control. The corresponding uncropped images for Figure 3 are shown on page 26 in the Supporting Information.
Characterization of P7 as an HDAC3-Directed PROTAC
The HDAC inhibitory potency of PROTAC P7 provided the best selectivity for HDAC3 over HDAC1/2, which was used as an argument to select P7 for further investigation. The concentration-dependence of P7-mediated HDAC3 degradation was also investigated in THP-1 cells using western blot. PROTAC P7 effectively induced HDAC3 degradation at nanomolar concentrations with a DC50 of 0.6 nM and a maximal degradation of about 90 % (Figure 4A, 4B and Figure S2A). Interestingly, an obvious hook effect was observed at a concentration≥3.3 μM. Notably, PROTAC P7 did not demonstrate a significant effect on the levels of HDAC1, HDAC2 and HDAC6, while significant effects on HDAC4 were observed at concentrations≥3.3 μM. (Figure 4A and 4 C).
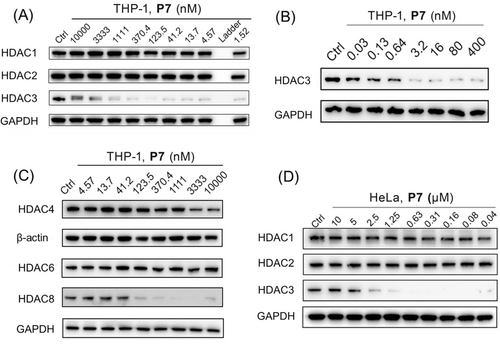
THP-1 cells were treated with the indicated concentrations of PROTAC P7 for 24 h. (A)The levels of HDAC1, 2, 3 (B) the level of HDAC3 and (C)the level of HDAC 4, 6 and 8 were detected using western blot. GAPDH and β-actin were used as a loading control. (D) HeLa cells were treated with the indicated concentrations of PROTAC P7 for 24 h. The levels of HDAC1, 2 and 3 were detected using western blot. GAPDH was used as a loading control. The corresponding uncropped images for Figure 4 are shown on page 27–28 in the Supporting Information.
We note that P7 can also trigger HDAC8 degradation, which provided a DC50 of 75 nM and Dmax of 94 % (Figure 4C and Figure S2B). To exclude the possible transcriptional effects by P7, quantitative reverse transcription polymerase chain reaction (RT-qPCR) assays were performed to examine the mRNA levels of HDAC3 and HDAC8. The HDAC3 mRNA level was significantly increased, whereas no significant effect was observed on the HDAC8 mRNA levels after P7 treatment for 24 h (Figure S2C and S2D), indicating that the cellular downregulation of HDAC3 and HDAC8 is due to direct protein degradation and not due to transcriptional downregulation. In addition, PROTAC P7 also effectively induced HDAC3 degradation in a dose-dependent manner in HeLa cells (Figure 4D). It is worth noting that although P7 triggered HDAC3 degradation in a dose-dependent manner in murine RAW264.7 cells (Figure S2E), its potency was much less than in THP-1 cells and HeLa cells, which can be attributed to differences between human and mice.
The ability of P7 to reduce HDAC3 levels in THP-1 cells was investigated further. The kinetics of HDAC3 degradation upon treatment with 0.5 μM of P7 demonstrated relatively rapid degradation. Degradation was observed after a 3-hours treatment, reaching about 95 % degradation after 48 h, which remains up to 72 h (Figure 5A). Next, the mechanism of HDAC3 degradation was investigated in both THP-1 and HeLa cells. We observed significant inhibition of HDAC3 degradation upon pre-treatment of HDAC3 inhibitor 16 or VHL ligand 2, which indicates that the ternary complex formation is required for HDAC3 degradation (Figure 5B). In addition, we also synthesized the enantiomeric control of P7 (NC-P7, Scheme S3), which is unable to recruit VHL E3 ligase. NC-P7 failed to induce HDAC3 degradation, indicating the essential role of VHL recruitment for degradation (Figure 5B). Similar results were also observed in HeLa cells (Figure 5C). Importantly, combined treatment with proteasome inhibitor bortezomib and P7 inhibited the HDAC3 degradation in HeLa cells (Figure 5C). Taken together, these results indicate that HDAC3 degradation induced by P7 depends on binding to both HDAC3 and VHL as well as proteasome activity.
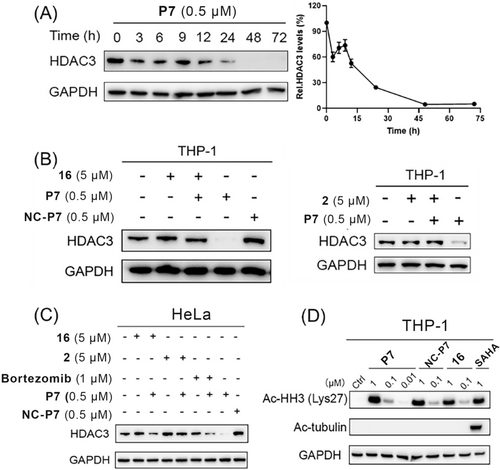
(A) Degradation kinetics of HDAC3 by PROTAC P7 in THP-1 cells. THP-1 cells were treated with 0.5 μM of PROTAC P7 for the indicated time. HDAC3 levels were detected using western blot. HDAC3 levels were quantified with Image J. Data are shown as mean ± SD, n=2 independent experiments. (B) Mechanistic investigation of HDAC3 degradation induced by PROTAC P7 in THP-1 cells. THP-1 cells were pre-treated with 5 μM of HDAC3 inhibitor 16 or VHL ligand 2 for 0.5 h, followed by 24 h treatment of PROTAC P7 at 0.5 μM. THP-1 cells were treated with 0.5 μM of NC-P7 for 24 h. HDAC3 levels were detected using western blot. GAPDH was used as a loading control. (C) Mechanistic investigation of HDAC3 degradation induced by PROTAC P7 in HeLa cells. HeLa cells were pre-treated with 5 μM of HDAC3 inhibitor 16 or VHL ligand 2 for 0.5 h, followed by 24 h treatment of PROTAC P7 at 0.5 μM or co-treated with bortezomib (1 μM) and PROTAC P7 (0.5 μM) for 24 h. HeLa cells were treated with 0.5 μM of NC-P7 for 24 h. HDAC3 levels were detected using western blot. GAPDH was used as a loading control. (D) THP-1 cells were treated with PROTAC P7, NC-P7, 16, and SAHA at indicated concentrations for 24 h. Ac-HH3 and Ac-tubulin levels were detected using western blot. GAPDH was used as a loading control. The corresponding uncropped images for Figure 5 are shown on page 28–29 in the Supporting Information.
We further investigated the effects of P7 on acetylated-histone H3 (Ac-HH3) as HDAC1/2/3 substrate and acetylated-tubulin (Ac-tubulin) HDAC6 substrate. The pan-HDAC inhibitor SAHA was used as a reference inhibitor. As shown in Figure 5D, compounds P7, NC-P7 and 16 could increase the intracellular level of Ac-HH3 in a dose-dependent manner, indicating their inhibition and/or degradation of HDAC1/2/3. However, the increased Ac-tubulin level was only observed in the SAHA-treated group, indicating that compounds P7, NC-P7 and 16 did not show significant effect on HDAC6 intracellularly. Taken together, we can conclude that P7 is a potent and selective HDAC3 PROTAC that can also trigger HDAC8 degradation at higher concentrations.
HDAC3 PROTAC P7 showed anti-inflammatory activity in THP-1 derived M1-like macrophages
After establishing P7 as an HDAC3-directed PROTAC, we compared the effect on inflammatory gene expression between HDAC3 degradation by P7 treatment, HDAC3 inhibition by 16 and treatment with negative control NC-P7. We employed LPS/IFNγ-stimulated THP-1 cells to monitor the expression of IL-10 as an anti-inflammatory gene and the expression of tumor necrosis factor α (TNFα), inducible nitric oxide synthase (iNOS), interleukin-6 (IL-6), interleukin-1β (IL-1β), and interleukin-12β (IL-12β) as pro-inflammatory genes. THP-1 cells can be polarized into M1-like phenotype in the presence of LPS/IFNγ and M2-like phenotype in the presence of IL-4. Firstly, the THP-1 cells were pre-treated with 0.3 μM of P7, NC-P7, 16 and 2 for 24 h, followed by treatment with LPS/IFNγ for another 24 h. Next, the corresponding gene transcription levels were measured by RT-qPCR. As shown in Figure 6, PROTAC P7 increased the expression of the anti-inflammatory gene IL-10 more than 10-fold compared to the vehicle control and decreased the expression of the pro-inflammatory gene IL-12β and iNOS. Although P7 slightly upregulated IL-6 and TNFα expression, its effect on IL-1β expression is limited. However, compound 16 and NC-P7 slightly increase the expression of IL-10 and showed no significant effect on pro-inflammatory genes, such as IL-12β, IL-6, TNFα and IL-1β. In contrast to NC-P7, compound 16 can significantly decrease iNOS expression probably owing to their different selectivity profiles for HDAC inhibition. It was worth noting that VHL ligand 2 did not show significant impact on both investigated anti-inflammatory and pro-inflammatory gene expression (Figure 6). Next, the IL-10 protein levels were assessed by an ELISA. As shown in Figure 7, PROTAC P7 significantly increased the IL-10 levels compared to NC-P7, 16 and vehicle control. Overall, these results indicate that HDAC degradation by PROTAC P7 provides more potent anti-inflammatory activity than HDAC inhibition by NC-P7 and 16 treatment in THP-1-derived cells in which an M1-like phenotype is induced.
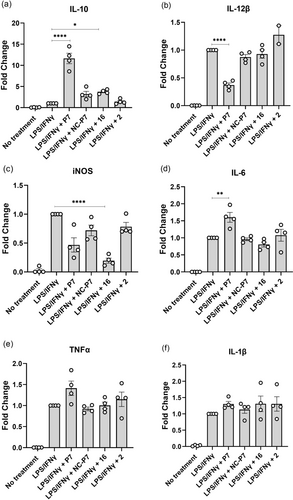
Effects of P7, NC-P7, 16 and VHL ligand 2 on pro- and anti-inflammatory gene expression of (a) IL-10, (b) IL-12β, (c) iNOS, (d) IL-6, (e) TNFα, (f) IL-1β in THP-1 cells. THP-1 cells were differentiated into macrophages with 10 ng/mL phorbol-12-myristate-13-acetate (PMA) for 24 h. Then, cells were treated with respective compounds at 0.3 μM for 24 h and stimulated with LPS/IFNγ for a further 24 h. Gene transcription was analyzed by RT-qPCR. Data are represented as mean ± SEM of 3 independent experiments. *P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001 vs vehicle group, one-way analysis of variance (ANOVA).
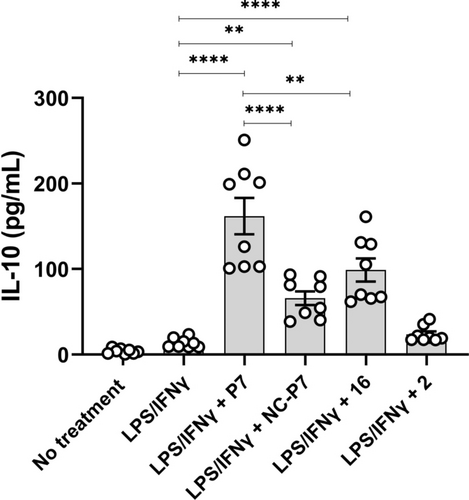
IL-10 protein levels were measured in the culture supernatants 24 h after LPS/IFNγ stimulation. Data are represented as mean ± SEM of 3–4 independent experiments. *P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001 vs vehicle group, one-way analysis of variance (ANOVA).
Subsequently, we investigated whether P7 can enhance the anti-inflammatory activity in IL-4-stimulated THP-1 cells (M2-like phenotype). THP-1 cells were pre-treated with 0.3 μM of P7 for 24 h, followed by treatment with IL-4 for another 24 h. However, RT-qPCR results demonstrated that P7 did not show significant impact on both anti- and pro-inflammatory gene expression (Figure S3). In addition, P7 had a similar impact on IL-10 levels as observed for NC-P7, 16 and vehicle control (Figure S4). Altogether, we conclude that HDAC3-directed PROTAC P7 demonstrated more potent anti-inflammatory activity than HDAC3-directed inhibitor NC-P7 and 16 in THP-1-derived M1-like macrophages by strongly increasing the level of anti-inflammatory cytokine IL-10.
HDAC3 PROTAC P7 demonstrated potent anti-inflammatory activity in human primary macrophages
Because P7 demonstrated potent anti-inflammatory activity in THP-1 cells with an M1-like phenotype, its anti-inflammatory potency was further investigated in a focused study on human macrophages from healthy donors. We firstly measured the HDAC3 degradation-inducing ability of P7 in primary macrophages that were derived from human peripheral blood mononuclear cells (PBMCs) from three healthy donors. western blot analysis showed that P7 efficiently triggered HDAC3 degradation in PBMC derived macrophages at both 0.15 μM and 0.3 μM, and more HDAC3 degradation was observed at 0.3 μM than 0.15 μM in D2 and D3 donors (Figure 8).

Human primary macrophages were treated with P7 at 0.15 μM and 0.3 μM for 24 h, respectively. HDAC3 levels were detected using western blot. GAPDH was used as a loading control. The corresponding uncropped images for Figure 8 are shown on page 30 in the Supporting Information.
Similar to THP-1 cells treatment, primary macrophages were polarized into an M1-like phenotype in the presence of LPS/IFNγ and an M2-like phenotype in the presence of IL-4. Primary macrophages were pre-treated with 0.3 μM of HDAC3 PROTAC P7 and HDAC3 inhibitor 16 for 24 h, followed by treatment with LPS/IFNγ for another 24 h. An ELISA assay showed that neither P7 nor 16 could significantly increase the IL-10 protein level (Figure 9a), which was not in line with the results in THP-1 derived M1-like macrophages. Nguyen et al. discovered that myeloid cell-specific HDAC3 knockout (MHD3KO) mice have lower levels of circulating TNFα and IL-6 compared to mice in which the HDAC3 catalytic activity was reduced due to mutations in the deacetylase-activating domain of nuclear receptor co-repressor 1/2 (NSDAD) mice and control mice.10 Therefore, we investigated the effects of P7 and 16 on the levels of TNFα and IL-6 via ELISA. As shown in Figure 9b and 9c, both P7 and 16 could significantly decrease the levels of TNFα and IL-6. It is worth noting that HDAC3-directed PROTAC P7 has much stronger ability to lower the levels of TNFα and IL-6 than HDAC3-directed inhibitor 16, indicating the potential of small molecule modulation of both the enzymatic and non-enzymatic HDAC3 functions in inflammation.

P7 regulated the cytokines secretion of (a) IL-10, (b) TNFα, and (c) IL-6. Human primary macrophages were treated with 0.3 μM of P7 and 16 for 24 h, followed by the treatment of LPS/IFNγ. The protein levels were measured in the culture supernatants 24 h after LPS/IFNγ stimulation using ELISA. Data are represented as mean ± SEM of 3 independent experiments. Statistical analyses were performed using the two-tailed Student's t-test. P values (* < 0.05, ** < 0.01, *** < 0.001, **** < 0.0001) are labeled in the Figures, and a P < 0.05 was considered to be statistically significant.
Given the critical role of M1 macrophages in initiating and sustaining inflammatory responses, we also investigated if HDAC3 PROTAC P7 could inhibit the polarization of M0-like macrophages towards an M1-like phenotype. The polarization status of macrophages was determined based on the following expression profiles: M1 (CD80 and CD86), and M2 (CD163 and CD206). As shown in Figure 10, P7 led to significant downregulation of CD68 and CD80, suggestive for the prevention of polarization towards a M1-like phenotype. Although no significant effects on the expression of CD11b and CD163, P7 induced upregulation of CD206, steering the polarization towards M0/M2-like phenotypes. Taken together, HDAC3 PROTAC P7 demonstrated more potent anti-inflammatory activity than HDAC3 inhibitor 16 in human primary macrophages by decreasing the pro-inflammatory cytokines (TNFα and IL-6), which can be explained by the observed inhibition of the polarization of M0-like macrophages towards M1-like phenotype.
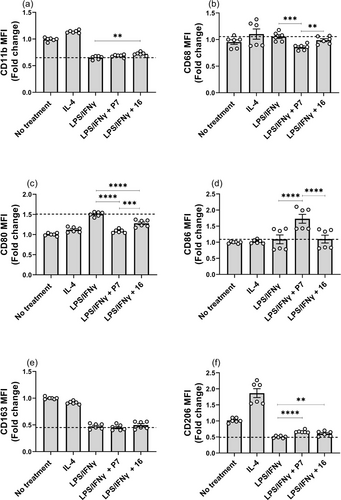
P7 blocked macrophage polarization from M0-like into M1-like. (a) CD11b, (b) CD68, (c) CD80, (d) CD86, (e) CD163, and (f) CD206. Human macrophages were either pre-treated with 0.3 μM of P7 and 16 for 24 h followed by the treatment of LPS/IFNγ. Data are represented as mean ± SEM of 3 independent experiments. Statistical analyses were performed using the two-tailed Student's t-test. P values (* < 0.05, ** < 0.01, *** < 0.001, **** < 0.0001) are labeled in the figures, and a P < 0.05 was considered to be statistically significant.
HDAC3 PROTAC P7 time-dependently demonstrated anti-inflammatory activity in human primary macrophages
Because HDAC3-directed PROTAC P7 showed more potent anti-inflammatory activity than HDAC3-directed inhibitor 16, the time-dependency of P7 was further explored. The human primary macrophages were either pre-treated with 0.3 μM of P7 for 24 h followed by the treatment of LPS/IFNγ [from now on referred to as LPS/IFNγ+P7 (Pre)] or simultaneously treated with 0.3 μM of P7 and LPS/IFNγ [from now on referred to as LPS/IFNγ+P7 (Simul)]. Firstly, RT-qPCR assays were performed to evaluate the effects on the expression of IL-10 as an anti-inflammatory gene and the expression of TNFα, IL-6, and IL-1β as pro-inflammatory genes. Of note, LPS/IFNγ+P7 (Pre) significantly downregulated IL-1β and IL-6 (Figure 11b and 11c). However, LPS/IFNγ+P7 (Simul) had no effect on the expression of IL-10, but significantly upregulated the expression of IL-6. In addition, LPS/IFNγ+P7 (Simul) resulted in decreased IL-1β and increased TNFα gene expression slightly (Figure 11b and 11c). Downregulation of IL-10 and upregulation of TNFα gene expression upon pretreatment with P7 were also significant but to a lesser extent than IL-1β and IL-6 (Figure 11a and 11d).
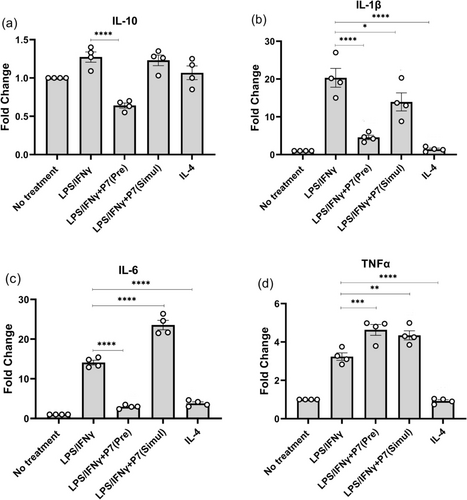
Time-dependent effects of P7 on pro- and anti-inflammatory gene expression of (a) IL-10, (b) IL-1β, (c) IL-6, and (d) TNFα in human macrophages. Human macrophages were either pre-treated with 0.3 μM of P7 for 24 h followed by the treatment of LPS/IFNγ [referred to as LPS/IFNγ+P7 (Pre)] or simultaneously treated with 0.3 μM of P7 and LPS/IFNγ [referred to as LPS/IFNγ+P7 (Simul)]. Gene transcription was analyzed by RT-qPCR. Data are represented as mean ± SEM of 3 independent experiments. Statistical analyses were performed using the two-tailed Student's t-test. P values (* < 0.05, ** < 0.01, *** < 0.001, **** < 0.0001) are labeled in the figures, and a P < 0.05 was considered to be statistically significant.
Mullican et al. reported that HDAC3 can lead to repression of M2 macrophage activation, and deletion of HDAC3 can augment M2 polarization in mouse macrophages.21 In this perspective, we aimed to investigate whether P7 can potentially enhance the anti-inflammatory ability of M2 macrophages in human macrophages. However, neither IL-4+P7 (Pre) nor IL-4+P7 (Simul) increased gene expression of IL-10 in IL-4 stimulated M2-like macrophages. Furthermore, no significant influence was observed in terms of the expression of IL-1β, IL-6, and TNFα (Figure S5).
Next to IL-10, TNFα, and IL-6, the secretion of other cytokines from human macrophages after P7 treatment was evaluated. As shown in Figure 12, the levels of classical pro-inflammatory cytokines TNFα and IL-6 were remarkably increased in macrophages after LPS/IFNγ exposure. Both LPS/IFNγ+P7 (Pre) and LPS/IFNγ+P7 (Simul) treatment significantly decreased the levels of IL-6 and TNFα. A similar decrease was observed in other pro-inflammatory cytokines after the treatment of P7, including interleukin-12p40 (IL-12p40) and interferon gamma-induced protein 10 (IP-10). Furthermore, the other pro-inflammatory cytokines i.e. interleukin-12p70 (IL-12p70), IL-1β, thymus- and activation-regulated chemokine (TARC), and interleukin-23 (IL-23) exhibited the same decreasing trend. It is worth noting that LPS/IFNγ+P7 (Pre) treatment showed stronger ability to lower pro-inflammatory cytokines than LPS/IFNγ+P7 (Simul) treatment. While both LPS/IFNγ+P7 (Pre) and LPS/IFNγ+P7 (Simul) treatments also attenuated the secretion of anti-inflammatory cytokines like IL-4 and IL-10 slightly. As for the IL-4-stimulated M2-like macrophages, IL-4+P7 (Pre) significantly decreased the level of IL-6 and moderately increased the level of IL-10, but there was no clear effect on the other cytokines (Figure S6). To summarize, while P7 did not significantly enhance the anti-inflammatory IL-4 activated macrophages, our results validated its strong ability to inhibit the pro-inflammatory genes and cytokines from LPS/IFNγ-stimulated human macrophages. These findings demonstrated that P7 treatment provided a remarkable anti-inflammatory activity in a time-dependent manner.
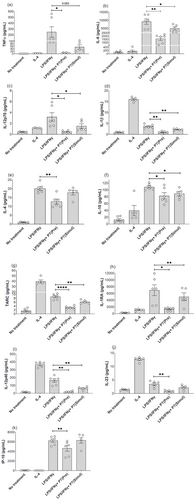
P7 time-dependently regulated the cytokines secretion of (a) TNFα, (b) IL-6, (c) IL-12p70, (d) IL-1β, (e) IL-4, (f) IL-10, (g) TARC, (h) IL-1RA, (i) IL-12p40, (j) IL-23, and (k) IP-10 in human macrophages. Data are represented as mean ± SEM of 3 independent experiments. Statistical analyses were performed using the two-tailed Student's t-test. P values (* < 0.05, ** < 0.01, *** < 0.001, **** < 0.0001) are labeled in the figures, and a P < 0.05 was considered to be statistically significant.
Next, we also investigated if P7 could time-dependently inhibit the polarization of M0-like macrophages towards an M1-like phenotype. Both LPS/IFNγ+P7 (Pre) and LPS/IFNγ+P7 (Simul) resulted in significant downregulation of CD68 and CD80, suggestive for the prevention of polarization towards a M1-like phenotype. In addition, LPS/IFNγ+P7 (Pre) induced upregulation of CD11b and CD206, steering the polarization towards M0/M2-like phenotypes (Figure 13 and Figure S7). Although there was no difference in the expression of CD163, both LPS/IFNγ+P7 (Pre) and LPS/IFNγ+P7 (Simul) caused a significant increase in CD86 (Figure 13 and Figure S7). Furthermore, a significant decrease in the expression of CD68 and an increase in the expression of CD206 were also observed in IL-4+P7(Pre)-treated macrophages compared to M2-like controls. Conversely, treatment with P7(Pre) resulted in the upregulation of CD86 expression and downregulation of CD163 expression of M2-like macrophages (Figure S8 and S9). Although the specific effect of P7 on M2-like macrophages remains uncertain, the combination of expression and protein levels of pro- and anti-inflammatory cytokine and phenotypic macrophage markers suggests that P7 can time-dependently inhibit the polarization of M0-like macrophages into M1-like macrophages in response to LPS/IFNγ stimulation.
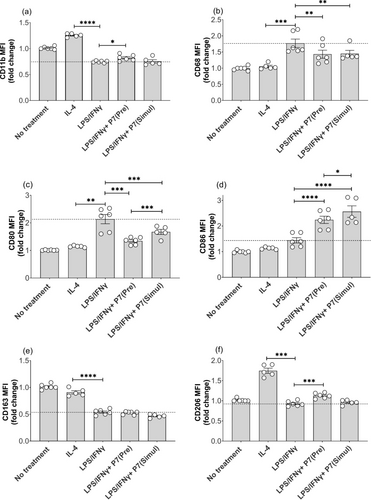
P7 time-dependently blocked macrophage polarization from M0-like into M1-like in a time-dependent manner. (a) CD11b, (b) CD68, (c) CD80, (d) CD86, (e) CD163, and (f) CD206. Human macrophages were either pre-treated with 0.3 μM of P7 for 24 h followed by the treatment of LPS/IFNγ [referred to as LPS/IFNγ+P7 (Pre)] or simultaneously treated with 0.3 μM of P7 and LPS/IFNγ [referred to as LPS/IFNγ+P7 (Simul)] Data are represented as mean ± SEM of 3 independent experiments. Statistical analyses were performed using the two-tailed Student's t-test. P values (* < 0.05, ** < 0.01, *** < 0.001, **** < 0.0001) are labeled in the figures, and a P < 0.05 was considered to be statistically significant.
Conclusion
Dysregulated functions of HDACs are deeply associated with various cancers, inflammatory diseases, and neurodegenerative disorders. The targeting of HDAC isoenzymes such as HDAC3 is gaining attention because of the potential to treat inflammatory diseases. Current insights indicate that the catalytic activity and the non-catalytic functions of HDAC3 play differential roles in inflammation. Therefore, we aimed to develop novel modalities to target the HDAC3 non-catalytic functions by using the PROTAC strategy.
In the past years, several HDAC3 PROTACs have been reported, which have however been only investigated as potential therapeutics against cancers. For example, the first reported HDAC3 PROTAC, XZ9002, triggered HDAC3 degradation (DC50=44 nM) in MDA-MB-468 cells and potently inhibited the proliferation of cancer cells.46 Smalley et al. reported another HDAC3 PROTAC, JPS036, which degraded HDAC3 in HCT116 cells with a DC50 value of 0.44 μM. Interestingly, JPS036 failed to induce apoptosis in HCT116 cells, in contrast to HDAC1/2 PROTACs.47 However, to address the well-described non-enzymatic functions of HDAC3, we developed a novel HDAC3-directed PROTAC, P7. This PROTAC connects an HDAC ligand to the VHL instead of the CRBN E3 ligase ligand that we previously explored.32 PROTAC P7 induced HDAC3 degradation in both THP-1 and HeLa cells, with a DC50 of 0.6 nM and Dmax of about 90 % in THP-1 cells, whereas HDAC8 was degraded at much higher concentrations and HDAC1, 2, 4, and 6 remained unaffected. Both RT-qPCR and ELISA results confirmed that P7 can enhance the expression of the anti-inflammatory cytokine IL-10, which is in line with the result of siRNA-mediated HDAC3 downregulation.20 These promising results in THP-1-derived macrophages raised our interest to investigate the effects of P7 on primary human macrophages. P7 triggered a decrease in HDAC3 levels at low micromolar concentrations in human primary macrophages. It is worth noting that P7 reduced the secretion of pro-inflammatory cytokines such as TNFα and IL-6 to a remarkably greater extent than HDAC3 inhibitor 16 did. This is in line with literature describing that MHD3KO mice have lower levels of circulating TNFα and IL-6 compared to NSDAD mice and control mice.10 Macrophage polarization is crucial in the process of inflammation. Of particular significance, P7 is able to prevent the polarization of M0- to M1-like macrophages in response to LPS/IFNγ stimulation in human primary macrophages. Overall, these results contribute to the evidence that HDAC3-directed PROTACs, such as P7, have the potential to suppress inflammation.
Altogether, we have developed a novel HDAC3-directed PROTAC that enables efficient and selective reduction of HDAC3 levels at concentrations in the low micromolar range. We firstly described the application of HDAC3 PROTACs beyond oncology into the field of inflammation. Application of an HDAC3 PROTAC in THP-1-derived macrophages showed a clear shift in the direction of anti-inflammatory cytokine expression, whereas application of this PROTAC in human primary macrophages led to a block of macrophage M1 polarization by decreasing the pro-inflammatory cytokines. Importantly, the HDAC3 PROTAC demonstrated much stronger anti-inflammatory potency than the corresponding inhibitor. In conclusion, both observations demonstrate the added value of the PROTAC strategy to reduce HDAC3 levels, which allows for interference with the non-catalytic functions of HDAC3. This indicates that HDAC3 PROTACs provide a small-molecule alternative for knock-out studies, and thus have great potential in pharmacology.
Acknowledgments
Chunlong Zhao, Shipeng Chen, Deng Chen, and Jianqiu Zhang are funded by China Scholarship Council (grant no. 202006220019 for Chunlong Zhao, grant no. 201808350110 for Shipeng Chen, grant no. 201907720019 for Deng Chen, grant no. 202107060008 for Jianqiu Zhang). We acknowledge the European Union for funding by the IN-ARMOR project (Project No. 101080889). We acknowledge Hangyu Zhou from Department of Chemical and Pharmaceutical Biology, Groningen Research Institute of Pharmacy (GRIP), University of Groningen for help on the purity test and Qing Chen from Department of Pathology and Medical Biology, University Medical Center Groningen, University of Groningen for the help with the experiments on primary macrophages.
Conflict of interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.