A Glycosidic-Bond-Based Mass-Spectrometry-Cleavable Cross-linker Enables In Vivo Cross-linking for Protein Complex Analysis
Graphical Abstract
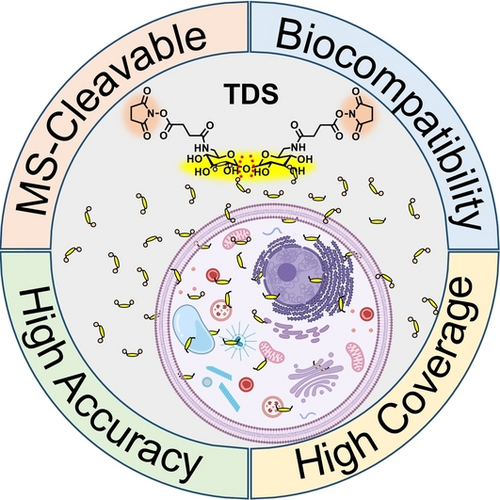
A glycosidic-bond-based mass-spectrometry-cleavable cross-linker has been developed that enables in vivo cross-linking for analyzing protein complexes. The MS-cleavable glycosidic bond of trehalose disuccinimidyl ester (TDS) improves the accuracy of identification while increasing the search throughput. In addition, in vivo cross-linking is enabled in the living cell microenvironment because of the high biocompatibility, bioorthogonality, and amphipathicity of TDS.
Abstract
Chemical cross-linking mass spectrometry (CXMS) has emerged as a powerful technology to analyze protein complexes. However, the progress of in vivo CXMS studies has been limited by cross-linking biocompatibility and data analysis. Herein, a glycosidic bond-based MS-cleavable cross-linker of trehalose disuccinimidyl ester (TDS) was designed and synthesized, which was fragmented in MS under CID/HCD to simplify the cross-linked peptides into conventional single peptides via selective cleavage between glycosidic and peptide bonds under individual MS collision energy. Consequently, the cross-linking identification accuracy and throughput were significantly enhanced, and the popular MS mode of stepped HCD was allowed. In addition, TDS showed proper cell-penetrating properties while being highly water-soluble, making it non-DMSO dependent during solubilization. Collectively, TDS provides a promising toolkit for CXMS characterization of living systems with high biocompatibility and accuracy.
Conflict of interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The mass spectrometric data were shared on iProx (https://www.iprox.org/), an official member of ProteomeXchange Consortium, and are publicly available as of the date of publication. Project ID: IPX0004418000.