Water Networks Repopulate Protein–Ligand Interfaces with Temperature
Graphical Abstract
Cryo-cooled water molecules block protein motions that impact protein–ligand interactions. Paired cryo and room-temperature comparisons of ligand-bound Hsp90α structures show that water networks and protein conformations are more mobile at room temperature. Revealing such alternate conformational states that are masked by cryo-cooling provides new insights towards drug discovery.
Abstract
High-resolution crystal structures highlight the importance of water networks in protein–ligand interactions. However, as these are typically determined at cryogenic temperature, resulting insights may be structurally precise but not biologically accurate. By collecting 10 matched room-temperature and cryogenic datasets of the biomedical target Hsp90α, we identified changes in water networks that impact protein conformations at the ligand binding interface. Water repositioning with temperature repopulates protein ensembles and ligand interactions. We introduce Flipper conformational barcodes to identify temperature-sensitive regions in electron density maps. This revealed that temperature-responsive states coincide with ligand-responsive regions and capture unique binding signatures that disappear upon cryo-cooling. Our results have implications for discovering Hsp90 selective ligands, and, more generally, for the utility of hidden protein and water conformations in drug discovery.
Introduction
Water is essential for protein structure and function.1 Water dynamics often determine how proteins adopt their global fold and how they transition between distinct functional states to catalyze enzymatic reactions.2-4 By energetically priming the binding sites for interactions, water networks enable differential ligand binding to related proteins.5, 6 Depending on differences in their position and energy, water molecules are either retained and reshuffled or displaced upon ligand binding.7 This information can be used productively in ligand discovery8 and is commonly explored computationally.9 Experimentally, the organization of water networks in proteins is typically studied using high-resolution X-ray crystal structures10 which are almost exclusively collected under cryogenic (cryo) conditions.11 While cryo-cooling has several benefits for sample stability,12 temperature changes also alter the protein energy landscape.13-15 Solvent mobility dominates atomic fluctuations above the glass transition temperature of ≈200 K.13 Below this temperature proteins undergo a dynamic transition to a glass-like, functionless solid.16 This provides a conceptual framework for how solvent mobility links to protein mobility and function. In fact the debated term “solvent slaving” is used to describe the relationship where protein conformational motions are slaved by water molecules in the hydration shell and bulk solvent.17 While it is clear that solvent plays essential roles in controlling important protein fluctuations,2 only few experimental studies investigate how temperature perturbs water structure instead of protein structure.5, 18, 19
The aim of this study is to link changes in water organization with temperature to modifications at the protein–ligand interface that impact the utility of structural data for ligand discovery. To demonstrate the importance of considering water network perturbations with temperature, we collected crystallographic data of five matched temperature pairs of molecular chaperone heat-shock protein 90 (Hsp90) to high-resolution both below (100 K) and above the glass transition temperature (room-temperature, RT). Since many tumor promoting proteins rely on Hsp90 for proper folding and activation, Hsp90 has been suggested as a promising therapeutic target.20 Yet, despite decades of efforts both in academia and industry, there are still no FDA approved drugs.21 To date, over 300 X-ray structures of the highly hydrated22 N-terminal ATP binding domain of Hsp90 alpha (Hsp90α-NTD) have been solved—all at cryogenic temperatures. We wondered whether cryo-cooling artifacts may have misled current ligand discovery efforts that have exclusively consulted structurally precise but possibly inaccurate cryogenic structures of Hsp90α. Our focus on water also resonates with the recent exploration of differential hydration patterns to develop Hsp90 isoform-selective inhibitors.23 Indeed, our results suggest that the sensitivity of water networks to cryo-cooling impacts protein structural ensembles and ligand binding interactions.
Results and Discussion
Temperature Alters Water-Mediated Protein–Ligand Interactions in Hsp90α
To investigate how temperature influences water organization at the ligand binding interface, we systematically compared X-ray crystal structures of Hsp90α solved with a series of ligands at both cryogenic temperature and RT. Comparing matched temperature pairs allowed us to isolate the impact of temperature changes on protein, ligand and water structure. To check whether ligand affinity affects active site plasticity, we compared two fragments to two clinical trial drug candidates (Figure S1). We selected adenine (ADE) and N-methyl-9H-purine-6-amine (N6M) as congeneric low-affinity fragments as they share the purine core of the native ATP ligand and differ by only 1 methyl group. The high-affinity drug-like compounds we picked are BIIB021 and EC144 as they share the purine core and a common substructure that is expanded by a 2-methylpent-4-yn-2-ol group in EC144, increasing both size and affinity (ki of 1.7 nM for BIIB021 and 0.2 nM for EC144).24, 25 To reduce bias, identical protocols were used to process diffraction data and refine structural models. Despite being solved at comparable resolutions ( =0.10±0.30 Å), RT structures had significantly lower average Rfree values (0.185±0.010) than cryogenic structures (0.212±0.221) (p<0.10; Supporting Information Tables S1–S2), suggesting higher quality models at RT. For example, the apo structures were solved at similar resolutions at both temperatures (ΔRes of 0.05 Å) while the Rfree value was 2.4 % better at RT compared to its cryogenic counterpart. On average, cryo-cooling crystals caused a 3 % (±2 %) reduction in unit cell volume (Figure S2); similar to previous studies.15, 26
=0.10±0.30 Å), RT structures had significantly lower average Rfree values (0.185±0.010) than cryogenic structures (0.212±0.221) (p<0.10; Supporting Information Tables S1–S2), suggesting higher quality models at RT. For example, the apo structures were solved at similar resolutions at both temperatures (ΔRes of 0.05 Å) while the Rfree value was 2.4 % better at RT compared to its cryogenic counterpart. On average, cryo-cooling crystals caused a 3 % (±2 %) reduction in unit cell volume (Figure S2); similar to previous studies.15, 26
To identify temperature-driven changes in local, water-mediated protein–ligand interactions, we used solvent accessible surface areas (SASA) to classify water molecules as exposed, buried or ligand-proximal (within 4.5 Å of ligand). On average, RT structures have significantly fewer buried, exposed, and buried ligand-associated waters (p<0.1; Supporting Information Table S1), as has been observed before.19 The position and number of waters near smaller fragment ligands are more consistent between temperatures than for larger drug-like molecules that coordinate more neighboring waters. For instance, despite the higher resolution at RT, several waters are lost in the structure of Hsp90α bound to the drug-like ligand BIIB021 which shifts by an RMSD of 0.58 Å compared to cryogenic temperature (Figure 1A). Considering all waters across all structures, we observed lower normalized B-factors of waters at cryogenic temperature than at RT (Table S3). Curiously, conserved buried ligand-associated waters have, on average, lower normalized B-factors at RT than cryogenic temperature (Supporting Information Figure S3 and Table S3), indicating decreased mobility despite increased thermal fluctuation at higher temperature while shifting on average 0.84 Å (±0.28 Å) compared to their cryogenic reference (Table S1).
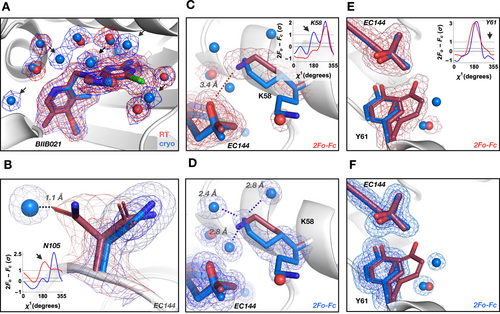
Temperature alters water-mediated protein–ligand interactions in Hsp90α. A) Water structure (shown as spheres) changes with temperature in the ligand binding site (BIIB021 shown with thick sticks). Several waters are not visible (arrows) at RT (red) or shift, despite higher resolution of the paired cryogenic (blue) dataset. B) A water molecule present in the Hsp90α+EC144 cryogenic structure restrains the conformational plasticity of Asn105 seen at RT; inset—Ringer plot. Water differences reorient charged residue Lys58 and change H-bonding pattern with the ligand EC144. Inset in (C)—Ringer plot indicates one side chain conformer at RT (red; C) and two at cryogenic temperature (blue; D). E) and F) Temperature alters water coordination at the unit cell interface (symmetry mate Tyr61) and changes ligand position. Inset—Ringer plot indicates two side chain conformers at RT (E) and one at cryogenic temperature (F). 2mFo-DFc maps are contoured at 1σ (red for RT, blue for cryogenic temperature) in panel (A) and contoured at 2σ (dark red/blue mesh) and 0.6σ (light red/blue mesh) for all other panels.
To reveal hidden changes in protein side chain conformations that coincide with temperature-induced alterations in water networks, we sampled electron density around side chain dihedral angles using Ringer.27 We observed two main effects of cryogenically frozen waters. First, well-defined cryogenic waters can suppress protein side chain conformations that are present at RT. For instance, Asn105 in Hsp90α+EC144 occupies two conformations at RT whereas, at cryo, a water molecule replaces one of the Asn105 RT conformers (Figure 1B). Second, temperature-driven differences in water coordination can create alternative hydrogen bonding patterns that reposition ligands. For instance, in the EC144 complex the ligand shifts by an RMSD of 0.77 Å depending on the temperature-tuned interaction with the side chain of Lys58. At RT, Lys58 forms a direct hydrogen bond at 3.4 Å with EC144 (Figure 1C), whereas at cryogenic temperature Lys58 is locked into interactions with three cryogenic waters away from the ligand (Figure 1D). The RT observation is consistent with a published 2.3 Å cryogenic structure of Hsp90β where Lys58 does not interact with EC144.24 Similar phenomena that lead to changes in ligand position are seen at the unit cell interface that typically compresses upon cryo-cooling. At RT the symmetry mate of Tyr61 from the neighboring unit cell is free to occupy two conformations (Figure 1E); at cryogenic temperature Tyr61 is restricted to a single conformation. Again, the alternative RT conformation of Tyr61 is superseded by a second ordered water at cryo (Figure 1F) causing a shift in the methylpentynol moiety of EC144 between cryogenic and RT structures.
Flipper Conformational Barcodes Report on Protein-Wide Changes in Side Chain Populations
To understand how widespread temperature-induced shifts in conformations are in these Hsp90α structures, we developed an algorithm called Flipper (Figure 2A). Flipper assesses conformational ensembles two ways. First, Flipper counts the number of conformations of each residue by detecting peaks in electron density maps. This extension of the Ringer algorithm27 allows us to determine whether a residue gains or loses a conformation with temperature or ligand binding (Figure 2B). The benefit of using the electron density maps is that we avoid any bias from errors in the structural models. Secondly, Flipper peak integration approximates the relative population of each conformation of a particular residue. This allows us to detect changes in the population of major and minor conformations with temperature or ligand binding (Figure 2B). After subtracting the number of peaks in cryogenic structures from their ligand-paired RT structure, we visualize Flipper results as “conformational barcodes” to provide a bird's eye view of temperature-sensitive regions in Hsp90α (Figure 2C). This highlights that many residues gain or lose conformations between temperatures and that such changes are often idiosyncratic for each temperature pair. Notably, these changes often occur near the ligand binding interface, such as Arg46 (Figure 2B) or along the ligand responsive ATP lid region (Figure S4). Also, several residues near the active site maintain the same number of conformations but the population of the major conformation switches between temperatures, such as for Ser50 in Hsp90α+EC144 (Figure 2D). Again, it appears that cryogenically restrained water molecules are liberated at RT and enable the flip in the Ser50 population. Together, these findings underscore that temperature-induced changes in water networks drive local changes in ligand binding sites that affect both ligand and side chain conformations.
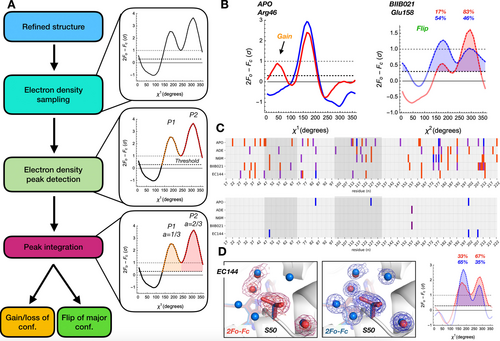
Flipper conformational barcodes report on changes in side chain populations by comparing electron density maps of otherwise identical structures at two temperatures. A) Schematic Flipper workflow of peak detection, integration, and classification of population shifts when comparing pairwise electron density maps determined at different temperatures across all residues. B) Examples of temperature shifts revealing a hidden alternative conformation (gain) in apo, and a change in a major conformation (flip) of BIIB021-bound Hps90α using Ringer where red and blue curves correspond to RT and cryo, respectively. C) Barcode plots showing (top) changes in number of side chain conformers based on χ1 for each temperature pair (2 additional peaks at cryogenic temperature in blue; 1 additional peak at cryogenic temperature in purple; 1 additional peak at RT in red) and (bottom) flips in major conformation between temperature pairs (χ1, blue; χ2, purple). Residues near active site are highlighted in dark grey. D) Active site proximal Ser50 shifts its major conformation between RT (left; red) and cryogenic temperature (center; blue) and is supported by relative peak areas of the Ringer plot (right; labelled as percentages). 2mFo-DFc maps are contoured at 2σ (dark red/blue mesh) and 0.6σ (light red/blue mesh) where red and blue corresponds to RT and cryogenic temperature, respectively.
Dynamic Regions Involved in Ligand Binding Respond to Temperature
In addition to local changes, we characterized temperature-induced global conformational changes in two ways. First, to understand how ligand-responsive regions differ with temperature, we calculated the backbone RMSDs for each model relative to the apo cryogenic temperature structure (Figure 3A). This revealed regions of the protein that change upon ligand binding, such as the characteristic lid repositioning that often occurs when larger ligands bind to the active site (Figure S4).28 Second, to isolate the effect of temperature on structure, we calculated RMSDs for each ligand temperature pair (Figure 3B). Several regions exhibit a noticeable response to temperature, including the ligand-responsive lid region of the binding site as well as several surface loops. Increased dynamics in these regions at RT is further supported by elevated normalized mainchain B-factors (Figure S5). The RMSD comparison of the BIIB021 temperature pair revealed that a loop in the back of the binding site lid occupies two distinct conformations at RT that differ by up to 2 Å from the single conformation we observe in our electron density map at cryogenic temperature (Figure 3B, C). To ensure that the lack of the second conformation did not originate from the lower resolution of our cryogenic temperature structure, we confirmed that a single conformation of this loop was seen in another cryogenic structure of BIIB021,29 solved at identical resolution as our RT structure (Supporting Information Figure S7). This temperature sensitivity is also seen for Leu122 and Glu123, which reposition their side chain conformations in response to temperature.
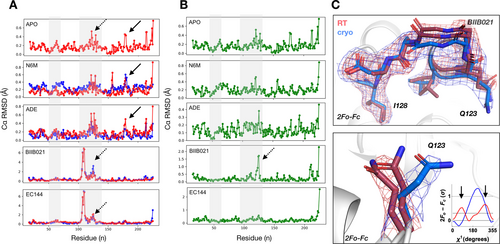
Dynamic regions involved in ligand binding respond to temperature. Comparison of backbone RMSDs of A) each structure (RT in red; cryogenic temperature in blue) against the cryogenic Hsp90α apo structure, and B) matched structures within each temperature pair highlights that regions responding to ligand binding are temperature sensitive (highlighted in grey). C, top) Electron density maps show multiple conformations and repositioning of an entire loop by up to 2 Å in the ligand binding lid region in BIIB021 bound structures upon shifting temperature (dashed arrows in A and B). C, bottom) Ringer plot showing that Glu123 is re-positioned by temperature and exhibits an additional conformation at RT (red) compared to cryogenic temperature (blue). 2mFo-DFc maps are contoured at 1σ (RT in red; cryogenic temperature in blue). Please note that the y-axis scaling is different between ligands in A and B. An unscaled version of Figure 3A is added to the Supporting Information as Figure S6.
Cryo-Cooling Idiosyncrasies Have a Larger Impact on Protein Structure than Ligand Binding
To determine whether temperature or ligand binding had a greater impact on Hsp90α structure, we compared side chain and backbone conformations between structures solved at the same temperature among either fragments or drug-like molecules. For example, for “cryo-drugs” we calculated the residue-based RMSD heatmaps of the BIIB021 and EC144 complexes both solved at cryogenic temperature (Figure 4A). Overall, both drug-like molecules and fragments show much smaller RMS deviations at RT than at cryo. RMSD heatmaps for drug-like molecules at RT and cryogenic temperature show agreement in the temperature-sensitive binding site lid with an RMSD of up to 1.5 Å (Figure 3). On the contrary, several high RMSD areas at cryo, for instance near the N- and C-terminus, show no signature at RT. The reduced noise level at RT reveals that the apparent hotspots in cryogenic temperature heatmaps are in fact idiosyncratic cooling artifacts possibly from variable cooling rates30 rather than due to differences in data quality (Supporting Information Table S4); the resolution of cryo-fragment structures are comparable (ΔRes≈0.1 Å). While there are no apparent signatures in the comparison of fragments to drug-like molecules at cryo, it is of note that the respective RT heatmaps reveal shared signatures even between small and large ligands both inside and outside the binding site (Figure 4A). Based on these RMSD heatmaps it appears that genuine differences and similarities that cannot be extracted from noisy cryogenic temperature comparisons can indeed be uncovered at RT. Mapping the drug-like RMSD comparison onto the structure of Hsp90α+EC144 at each temperature shows that regions with higher variability at cryogenic temperature impact not only the surface loops but also regions adjacent to the ligand binding site (Figure 4C and D). In addition, conformational barcode plots further illustrate unique temperature induced conformational signatures (Figure 4B). Considering a total of about 210 residues, barcodes of drug-like molecules show agreement for 8 residues, and differences for 15 residues between RT and cryo; fragment barcodes indicate agreement for 6 residues and differences for 18 residues between temperatures. Overall, these data suggest that noise in cryo datasets masks genuine differences of related ligands that can be revealed when comparing data obtained at RT.
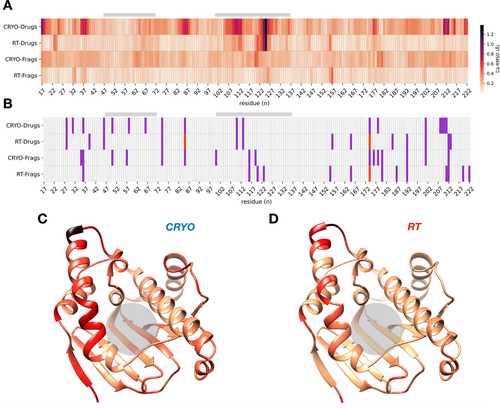
Idiosyncrasies of cryo-cooling have a larger impact on protein structure than ligand binding in RMSD heatmaps and conformational barcode plots. A) Heatmap of backbone RMSDs between drugs and fragments at the same temperature distinguishes genuine differences from noisy cryogenic artifacts (RMSD gradient colored from beige to black from 0 to 1.5 Å). B) Barcode plot of the absolute difference in the number of ringer peaks (1, purple; 2, red) between drug-like molecules (BIIB021 vs EC144) and fragments (NM7 vs ADE) collected at the same temperature. C and D) RMSD values with color gradient from (A) plotted onto ribbon diagram of Hsp90α+EC144 colored by “cryo-drugs” (C) and “RT-drugs” (D). The ligand binding site is indicated by grey bars in (A) and (B) and grey circle in (C) and (D).
Conclusion
This work highlights how differences in water reorganization with temperature change the population of conformational ensembles in Hsp90α ligand complexes. Three important implications for exploring protein–ligand interactions emerge. First, cryogenically frozen water molecules mask alternative side chain and loop conformations that are seen at RT where water structure is more dynamic. Second, dynamic regions, such as those involved in ligand binding, are exquisitely temperature-sensitive. As cryo water engages binding site residues, ligands shift positions and change interaction patterns. Third, noise in cryo datasets has a larger impact on Hsp90α structure than ligand binding and hinders the identification of genuine orthosteric and allosteric signatures of ligand interactions.
This study extends previous work on the structural impact of crystallographic data collection temperature11, 31 by highlighting the involvement of water molecules in many changes in protein and ligand conformations. Similar to water restructuring upon protein mutation32 the thermodynamic features of the binding site that are considered for ligand discovery are altered via temperature-induced changes of binding site water networks. In line with different energetic penalties of displacing water at lower temperatures, we saw several examples of alternative side chain and loop conformations disappearing upon cryo-cooling. This is consistent with lower enthalpy and entropy states being favored at low temperatures.33 As these changes do not spare the dynamic binding site, ligand interactions and positions may be modified upon cooling. We have seen this, for instance, in the lost interaction between Lys58 and EC144 at cryo. We introduced the Flipper algorithm to further interrogate electron density maps for conformational gains, losses and flips. Flipper revealed that temperature-sensitive regions occur across all Hsp90α structure pairs, and affect ligand-free as well as liganded structures bound to fragments and drug-like molecules differently, even for similar ligands. We expected tightly binding ligands to be less affected by cryo artifacts than weakly binding fragments. However, we observed that drug-like molecules were remarkably susceptible to temperature-induced changes in water networks for example in the EC144 complex with a ki of 200 pM. This is counterintuitive since binding opposes motion and vice versa. Thus, it emphasizes unappreciated energetic penalties of water at protein–ligand interfaces, especially in highly hydrated binding sites like the ATP site in Hsp90α.3, 22, 34
Notably, cryo-cooling crystals increases the number of observable waters in electron density maps (Table S1). This creates the impression that RT maps are missing fundamental features. To illustrate why this is deceptive, let us borrow the analogy of the formation of an icicle. Cold temperatures capture an image of dripping water that is invisible at room temperature. However, while more water is captured in the resulting icicle, it fails to reflect the dynamic nature of the event, nor is the representation of dripping water realistic, despite the striking image it creates. Back to our electron density maps, lack of order does not imply lack of function; important structural elements are often invisible in electron density maps of partially or entirely disordered proteins. Yet, statistically significant differences in Rfree values suggest that RT models are generally of higher quality than cryo datasets at comparable resolutions—despite fewer ordered RT waters (Table S1).
With a view towards ligand discovery, we can leverage Flipper categories of conformation gains, losses and flips for computational docking. Flips in the major side chain conformation with temperature are particularly noteworthy, as standard protocols to prepare a structure for docking discard all minor conformations. Consequently, flipped residues often point in the opposite direction at cryo, as we have seen for Glu158 in the BIIB021 complex, and Ser50 and Asn105 in the EC144 complex (Figure 1 and 2). RT data can provide an alternative docking template with different steric and interaction patterns that will prioritize other molecules than at cryo.35 In addition, charged residue flips will change the electrostatic nature of the binding site considerably, as we have seen for Lys58 in the EC144 complex and Arg46 in the apo structure (Figure 1 and 2). However, even more subtle docking approaches that include residue flexibility will be affected by minor temperature-dependent residue changes revealed by Flipper. For instance, experimental occupancies of minor conformational states that are revealed only at RT can be used directly in docking as Boltzmann-weighted energy penalties.36 As we have shown before, this can reshuffle hit lists and lead to ligands with new chemical and physical properties that cannot be discovered otherwise. Finally, while water is key for ligand binding,5 water molecules are almost universally discarded in docking, with some notable exceptions.5, 37 But even for those few algorithms that consider penalties of water displacement upon ligand binding, water positions are either taken from cryo structures or computational simulations.9, 38 With the stark differences in the quantity and quality of water molecules at RT, considering RT water networks instead will re-prioritize ligands from virtual screens. Such expansions of chemical space are an interesting prospect for Hsp90 drug discovery, where, despite decades of research, still only few chemotypes are represented in clinical trials and none have reached FDA approval.39 New chemistry opens new avenues40 to address current side effects and other problems like the induction of the heat shock response.41
Our data suggest that non-systematic differences often do not reflect genuine responses to ligand binding but rather cryo-cooling idiosyncrasies. These often originate from differences in cooling rates as crystals of different sizes traverse a changing layer of liquid nitrogen.30 Consequently, widespread cryogenic artifacts hide and distort unique signatures of ligand binding and misinform ligand discovery and design. Our conformational barcode plots and heatmap signatures address this issue by revealing conformational signals in RT datasets that lie hidden in the noise of heterogeneous cryogenic datasets. Understanding these slight differences may provide important clues on how protein conformational changes in response to congeneric ligand series may be exploited to achieve selectivity among highly similar isoforms. More generally the results highlight that water reorganization with temperature widely modulates protein and ligand interactions through distortions of the conformational landscape one seeks to explore in ligand discovery.
Acknowledgements
This work was supported by ALSAC and 1R35GM142772-01 (to M.F.) and an Academic Programs Special fellowship (to T.R.S.). We thank the High-Performance Computing Facility, Protein Production Facility, St. Jude X-ray Center, the SERCAT beamline staff for ongoing support and C. M. Fischer for proof-reading. FMX and AMX beamlines are primarily supported by the National Institutes of Health, National Institute of General Medical Sciences (NIGMS) through a Center Core P30 Grant (P30GM133893), and by the DOE Office of Biological and Environmental Research (KP1607011). As part of NSLS-II, a national user facility at Brookhaven National Laboratory, work performed at the CBMS is supported in part by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences Program under contract number and DE-SC0012704.
Conflict of interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
Structures have been deposited to the Protein Data Bank under following accession codes: 7S8Y, 7S8Z, 7S90, 7S95, 7S98, 7S99, 7S9F, 7S9G, 7S9H, 7S9I. The Flipper algorithm is openly available on github at https://github.com/TheFischerLab/Flipper.