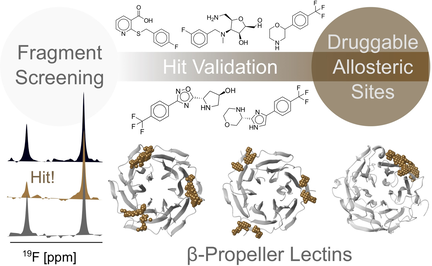
Druggable Allosteric Sites in β-Propeller Lectins
Graphical Abstract
Drug-like inhibitors for bacterial carbohydrate-binding proteins called lectins are challenging. Computational and experimental analyses (NMR and SPR) of related β-propeller lectins from Burkholderia ambifaria (BambL), Ralstonia solanacearum (RSL), and Aspergillus fumigatus (AFL) identified druggable secondary pockets, which will support the design of allosteric inhibitors as new therapeutics against antibiotic-resistant pathogens.
Abstract
Carbohydrate-binding proteins (lectins) are auspicious targets in drug discovery to combat antimicrobial resistance; however, their non-carbohydrate drug-like inhibitors are still unavailable. Here, we present a druggable pocket in a β-propeller lectin BambL from Burkholderia ambifaria as a potential target for allosteric inhibitors. This site was identified employing 19F NMR fragment screening and a computational pocket prediction algorithm SiteMap. The structure–activity relationship study revealed the most promising fragment with a dissociation constant of 0.3±0.1 mM and a ligand efficiency of 0.3 kcal mol−1 HA−1 that affected the orthosteric site. This effect was substantiated by site-directed mutagenesis in the orthosteric and secondary pockets. Future drug-discovery campaigns that aim to develop small molecule inhibitors can benefit from allosteric sites in lectins as a new therapeutic approach against antibiotic-resistant pathogens.
Introduction
Bacterial infections, especially those involving biofilm formation, are becoming increasingly difficult to treat as antibiotic resistance is rising worldwide. Therefore, identifying new protein targets and anti-adhesives is required. Since carbohydrate-binding proteins (lectins) are found in many pathogenic microorganisms and involved in host recognition, adhesion and biofilm formation, targeting lectins evolved as an attractive strategy to treat bacterial and fungal infections.1
Lectins from pathogens often display a high affinity for mammalian carbohydrates, likely deriving from co-evolution.2 Thus, bacteria take advantage of these interactions to adhere and infect the host. A well-known example is the β-propeller lectin BambL from the Gram-negative bacterium Burkholderia ambifaria.3 This opportunistic pathogen belongs to a group of closely related bacterial strains, the Burkholderia cepacia complex, causing chronic infections and exhibiting multidrug antibiotic resistance. B. ambifaria affects immunocompromised patients as well as those suffering from cystic fibrosis (CF) and can cause pneumonia, respiratory failure and bacteremia.4 Moreover, B. ambifaria can promote sporadic outbreaks, but its epidemiology remains elusive.5 Several studies point to an underestimated role of BambL in affecting host cellular processes, which go beyond an adhesion to the human lung epithelium.6 Therefore, blocking the carbohydrate–BambL interactions is a potential avenue to treat chronic infections, but strategies for design of inhibitors are required.
The crystal structure of BambL revealed that the protein consists of two similar domains and trimerizes to form a 6-bladed β-propeller with 6 fucose-binding sites.3 Bacterial and fungal β-propeller is an efficient carbohydrate-binding fold presenting all binding sites on one face of the donut shape.7 In recent years, several inhibitors for BambL have been reported. Given the strong affinity of BambL for α-l-fucosylated monosaccharides (methyl α-l-fucopyranoside (MeFuc), Kd=1 μM) and complex carbohydrates (H type 2 tetrasaccharide, Kd=7.5 μM), the design of inhibitors has been focused on using carbohydrates as a starting point.3 Indeed, this approach has yielded potent BambL monovalent aryl-α-O-fucoside inhibitors with an affinity comparable to MeFuc.8 Moreover, multivalent compounds with 4 to 6 fucose or aryl-α-O-fucosyl analogues improved the selectivity and the affinity towards BambL with Kd ranging between 10 to 80 nM.8a, 9 However, the main limitation of such complex carbohydrate-based inhibitors is their molecular size, which limits their oral bioavailability and thus, complicates the future clinical approval.10 Consequently, small and orally bioavailable drug-like molecules targeting lectins from pathogens are highly desired, but challenging.
Lectins have been associated with a low druggability index due to their hydrophilic and solvent-exposed carbohydrate-binding sites.10, 11 To overcome these limitations, we have previously explored the concept of allosteric modulators for mammalian lectins.12 Allosteric modulators do not bind to the orthosteric (carbohydrate)-binding site, but target an alternative (allosteric) pocket that affects the orthosteric site and vice versa. Several druggable, allosteric pockets have been discovered for the mammalian lectins as DC-SIGN (CD209),13 including intra-domain allosteric network that modulates Ca2+ affinity of Langerin (CD207). This was followed by design of allosteric inhibitors for Langerin supporting the allosteric communication in mammalian lectins.12a, 14 Altogether, these discoveries paved the way for further search of potential allosteric pockets in lectins.
Motivated by previous reports, we assessed the druggability of β-propeller lectins with the focus on a bacterial lectin BambL. Competitive 19F and T2-filtered (CPMG) NMR allowed us to distinguish drug-like fragments binding to lectins in the orthosteric or the secondary sites. The hits were counter-screened by surface plasmon resonance spectroscopy (SPR) and protein-observed 1H–15N TROSY NMR (hereafter, TROSY NMR). The affinity and potential modulatory properties of the most promising fragments were determined in three orthogonal NMR experiments (TROSY, PrOF15 and 19F R2-filtered NMR). Finally, computational analysis was applied to predict druggable secondary sites in BambL and validated experimentally by site-directed mutagenesis and NMR.
Results and Discussion
Fragment Screening Against β-Propeller Lectins
Ligand-observed 19F and T2-filtered (CPMG) NMR are key methods for detection of weak fragment–protein interaction.16 This is owing to the T2 relaxation of the 19F nucleus, which is highly sensitive to the molecular tumbling changes of the small molecules in the unbound and protein-bound states.17 Therefore, 19F NMR screening of fragment mixtures is frequently used in drug discovery to estimate the protein druggability. Previously, we successfully applied our diversity-oriented fragment library and 19F NMR to discover drug-like molecules for mammalian lectins.11, 12, 13, 18
Encouraged by this discovery, we applied this approach to assess the druggability of BambL and related β-propeller lectins RSL and AFL from the bacterium R. solanacearum and the fungus A. fumigatus (Figure 1 a). These lectins have sequence and structure similarities with BambL (RSL: 76 % sequence identity, RMSD=0.56 Å, AFL: 39 % sequence identity, RMSD=1.84 Å). Similar to BambL, AFL and RSL have a low micromolar affinity for terminal α-l-fucose on animal and plant carbohydrates with the affinity (Kd) of 76.4 μM and 0.64 μM, respectively.19
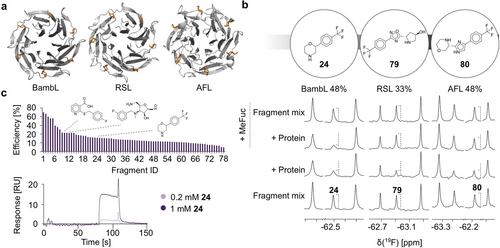
Druggability assessment of β-propeller lectins. (a) Cartoon representations of the crystal structures of the β-propeller lectins. Shown are BambL in complex with l-fucose (orange, PDB ID: 3ZZV), RSL (PDB ID: 3ZI8) and AFL (PDB ID: 4AGI). (b) CPMG NMR spectra of fragment mixtures containing 0.05 mM 24, 79 or 80 show a strong line broadening effect of 19F resonances (dashed line) in presence of 20 μM BambL/AFL and 40 μM RSL, whereas only 79 and 80 were competed with 10 mM MeFuc. (c) Shown are the structures of 19F NMR screening hits for BambL confirmed in SPR and TROSY NMR among 78 hits verified in SPR (top). SPR sensorgrams of the binding of 24 to BambL at two doses 0.2 mM and 1 mM (bottom).
To assess the druggability of β-propeller lectins, 350 fluorinated fragments were screened in 19F and CPMG NMR. Herein, we carefully monitored the chemical shift perturbations (CSPs) or the changes in peak intensity of 19F resonances. Fragments with CSP>0.01 ppm or peak reduction of 25–50 % were defined as “high” and “low” confidence 19F hits, respectively. In the CPMG NMR spectra, the changes in peak intensity of 20–50 % or more than 50 % were defined as “low” and “high” confidence CPMG hits, respectively. Only compounds fulfilling one of three criteria: 1) 19F hit only, 2) 19F and CPMG hit and 3) “high” confidence CPMG hit, were followed up in the counter-screening. As an example, Figure 1 b shows the identification of the fragment 24 bound to BambL in CPMG NMR experiment. Such fragments were used to derive a total hit rate. Interestingly, β-propeller lectins showed unusually high hit rates, that is, 33 % and 48 % for RSL and AFL/BambL, respectively. Such high total hit rates in a fragment screening originated either from a large number of the frequent-hitters (FHs) or the presence of several potentially druggable secondary sites.20 However, the FHs contribution to the total hit rates is likely low since previous studies identifying 10–15 % hit rates with the same library against C-type lectins found most hits to be specific.11, 13, 18b To further narrow down the number of potential hits, we defined the fragments targeting the orthosteric site using 10 mM MeFuc as a competitor. Surprisingly, only 2 “low” confidence fragments (<1 %) were identified for the orthosteric site of BambL, whereas 17 % and 5 % of fully or partially competed fragments were observed for AFL and RSL, respectively (fragments 79 and 80, Figure 1 b).
To unravel the potential fragment binding sites in β-propeller lectins, we applied a computational pocket prediction algorithm SiteMap.21 SiteMap identified three secondary pockets in the crystal structures of BambL in complex with α-l-fucose (PDB ID: 3ZW0, Figure S1) or H type 2 tetrasaccharide (PDB ID: 3ZZV),3 as well as in RSL (apo and holo, Figure S2 a,c). Further, one druggable site with a different shape and size was identified in AFL (Figure S3). In contrast, the predicted sites in BambL and RSL were located in the same areas with slightly differing shapes and sizes of the pockets, which was due to the differences in residues in the sites (Figure S2 b,d).
Taken together, high hit rates in 19F NMR and SiteMap computational pocket prediction analysis strongly suggest the availability of druggable pockets capable of accommodating drug-like molecules in β-propeller lectins.
Druggable Secondary Sites in BambL
To explore the concept of druggable secondary sites in β-propeller lectins, we chose BambL as an example. Given a large number of 19F NMR hits, we focused on 111 fragments with the strongest effects in 19F and CPMG NMR (Figure S4 a). Thereby, 13 compounds were removed due to their poor solubility resulting in 98 hits subjected to the orthogonal screening using SPR and TROSY NMR. Briefly, SPR confirmed a dose-dependent interaction of 78 out of 91 fragments with BambL (Figures 1 c and S4 b). To further narrow the number of hits, TROSY NMR as a “gold standard” for hit validation was applied.22 For this, we performed a partial protein backbone assignment using site-directed mutagenesis as described in the Supporting information. Further, we confirmed 10 out of 39 compounds with the strongest effects in SPR, 19F and TROSY NMR (Figure S4 c) and assessed these in TROSY NMR for a dose-dependent binding, which identified compounds 10, 12 and 24 (Figure S5 a). Interestingly, the CSPs did not follow a single vector suggesting a binding mechanism other than a one-site binding model, which however was used to derive their affinities (Kd) and ligand efficiencies (LE) as an approximation.23 The fragment 24 showed a two-fold stronger affinity (Kd=0.4±0.2 mM, Figure S5 b) and a better LE value of 0.29 kcal mol−1 HA−1 compared to 10 (Kd=0.8±0.4 mM, LE=0.23) and 12 (Kd=0.9±0.3 mM, LE=0.21), which was probably due to its smaller molecular weight. To estimate whether 24 serves as a good starting point for lead development, we verified its interaction with BambL in an orthogonal ligand-observed 19F R2-filtered NMR assay (Figure S6), which revealed the similar range Kd value to that obtained by protein-observed TROSY NMR (Kd=0.3±0.1 mM, LE=0.3 kcal mol−1 HA−1).
In parallel, we investigated whether the predicted secondary sites in BambL could host the fragments 10, 12 and 24. As SiteMap identified three druggable sites in BambL, we noticed only slight differences in the predicted sites of the crystal structure (PDB: 3ZW0). As an example, K23 (in three sites) and L87 (in one site) illustrate the differences in side chain orientation, which slightly changes the shape and the size of the predicted sites. Likely, these were only due to differences in side chain orientation and thus, were expected to be identical in solution. Consequently, we selected only one of the sites for the docking study, which is located at the interface between the monomers near the C-terminus forming narrow channels (T18, N20, K23, T25, G67, T69, G86 and L87, Figures 2 a and S7). Moreover, the hydrophilic residues surrounding the pocket make it suitable to accommodate ligands with polar groups. Indeed, we successfully docked these fragments using Glide (v. 7.8) in the predicted site (Figures 2 b and S8). The docking study suggested six residues (T18, K23, T25, G67, Y84 and L87) to play key roles in fragment binding. In particular, 24 bound to the predicted site with nearly identical pose indicating only a minor difference in orientation of morpholine ring in multiple binding poses (Figure S9). Likewise, 12 and 10 were also accommodated in the site showing H-bond interactions with the identified key residues.
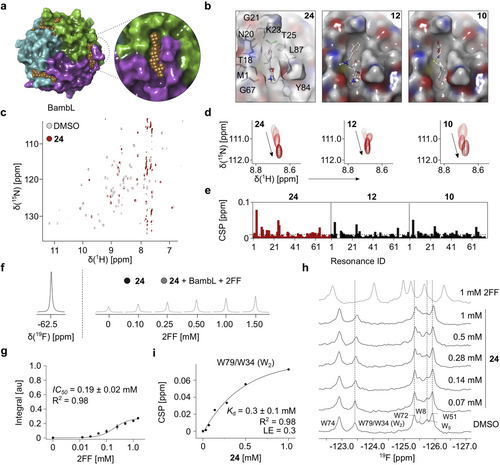
Identification of the druggable secondary sites in BambL. (a) BambL harbors three potential druggable binding sites, whereas only one secondary site (enlarged view) can accommodate drug-like molecules 24, 12 and 10 as predicted by SiteMap (PDB ID: 3ZW0). (b) Docking poses of 24, 12 and 10. (c) TROSY NMR of 15N BambL with DMSO or 24. (d) Shown is an example of dose-dependent CSPs upon addition of 24, 12 and 10. (e) CSP plots 24, 12 and 10 demonstrate that fragments perturbed similar resonances in 15N BambL, whereas 24 showed a larger magnitude of CSPs compared to 12 and 10. Dashed line indicates CSPs >0.01 ppm. (f),(g) Competitive T2-filtered 19F NMR yielded IC50 value of 2FF in presence of 1 mM 24 and 0.1 mM BambL. Notably, 2FF competed 24 only partially suggesting 24 bound to BambL distantly from the orthosteric site. (h) PrOF NMR of 0.1 mM 5FW BambL shows CSPs of all six 5FW resonances in presence of 1 mM 2FF. Moreover, 24 perturbed W79/W34 (W2 and W5, unassigned), W72 and W51 demonstrating an effect of remote site binders on the carbohydrate-binding site. (i) One-site fit of PrOF NMR titration data. The CSPs of W79/W34 (W2) upon addition of 24 were followed up to derive the affinity.
To support this prediction experimentally, we quantified the chemical shift perturbations in TROSY NMR spectra of 15N BambL in presence of fragments 10, 12 and 24. Despite only a partial protein backbone assignment, we observed that 10, 12 and 24 perturbed the same resonances in 15N BambL. This suggested that fragments targeted identical binding site in BambL, which supports our computational data (Figure 2 c–e).
Next, we confirmed that 24 bound to a secondary pocket distinct from the orthosteric site. For this, we employed a competitive ligand-observed 19F CPMG NMR using 24 as a fluorinated reporter and 2-deoxy-2-fluoro-l-fucose (2FF) as a competitor (Figure 2 f). Compared to MeFuc, 2FF binds to the orthosteric site of BambL with a slightly weaker affinity (Kd=18.8±2.3 μM24) and thus, it could have a better chance in competing 24. Indeed, 2FF demonstrated a dose-dependent binding to BambL with IC50 value of 0.19±0.02 mM being 10-fold weaker than reported previously (IC50=0.02 mM,24 Figure 2 g). Given that 2FF displaced 24 only partially, we concluded that 24 bound BambL in the secondary pocket.
To investigate the impact of 24 binding on the orthosteric site of BambL, we employed protein-observed 19F (PrOF) NMR.15 Previously, this method proved to be valuable for identification of small molecules targeting a bacterial lectin LecA.25 Given that BambL monomer contains six tryptophan residues in the carbohydrate-binding site (Figure S10 a), we sought to apply PrOF NMR to verify the impact of fragment binding on the orthosteric site. For this, we substituted tryptophan residues in BambL with 5-fluorotryptophanes (5FW) and assigned four out of six 5FW by site-directed mutagenesis (Figure S10 b). Next, we confirmed the activity of 5FW BambL using MeFuc and 2FF, where all six 5FWs showed a slow exchange on the NMR time scale (Figure S10 c,d). This demonstrated that both natural ligands are strong binders. Thus, PrOF NMR was not well suitable to determine its affinities hampering the accurate derivation of the Kd values (2FF: Kd=46±11 μM compared to reported Kd=18.8±2.3 μM,24 Figure S10 e). This is not surprising, given that similar limitations were reported for protein-observed 1H–15N HSQC NMR.26 However, PrOF NMR verified the impact of fragment (10, 12 and 24) binding on the orthosteric site of 5FW BambL through W51, W72 and W79/W34 (W2 and W5, Figure S11, Table S1). Moreover, titration of 24 to 5FW BambL perturbed 5FWs in a dose-dependent manner delivering a Kd of 0.3±0.1 mM in agreement with our previous results (Figure 2 h,i). Therefore, we investigated whether fragments could inhibit 5FW BambL interaction with 2FF. Notably, 24 remained bound to 5FW BambL in the presence of 2FF (Figure S10 f), as well as 12 (Figure S10 g). However, 24 did not inhibit 2FF-5FW BambL interaction in this assay (Kd=52±3 μM, Figure S10 h), which contradicted with our competitive 19F CPMG NMR result and thus, the modulatory properties of 24 remain to be proven.
Taken together, computational and experimental analyses using 19F CPMG NMR confirmed the presence of druggable secondary sites in BambL. Despite the inconclusive result about the inhibitory properties of 24, PrOF NMR demonstrated that binding of fragments 10, 12 and 24 influenced the orthosteric site in BambL, which strongly suggested the presence of a communication between the orthosteric and predicted sites. Given this, 24 was subjected to further studies.
Structure–Activity Relationship Study of 24
The aim of the SAR study was to improve the affinity of 24 using commercially available analogues (Table S2, Figure 3 a). For this, we employed computational and experimental TROSY NMR analyses. Briefly, we performed TROSY NMR of 16 analogues of 24 (Figure S12), which revealed the importance of the morpholine group in 24 given a fully and partially abrogated binding upon its replacement with piperidine (84), morpholine-3-one (91) and tetrahydro-2H-pyran-4-ol (92) groups, respectively (Figure S13 a). Notably, further modification on the amine group to 4-(2-aminoethyl)morpholine (90) was tolerated unlike a more hydrophobic and bulky change as 5-bromopyrimidine (89, Figure S13 b). Moreover, we observed that the replacement (95) or lack (96) of CF3 and changing the position of the benzyl group from 1 (96) to 2 (87) did not abrogate BambL binding (data not shown). Therefore, we explored the role of the benzyl group by changing it to 1,3-dichlorobenzene (86), 3-methylpyrazole (88), methyl acetate (98), N-formylpiperidine (97), tetrazole (83), 2-bromo- (85) or thiophene (94). As result, analogues 83, 85, 86, 88, and 94 preserved binding to 15N BambL (Figures S14 a and 3 b). As only 83 promoted more total CSPs above 0.01 ppm in 15N BambL compared to 24, we confirmed it in a competitive 19F CPMG NMR (Figure 3 c). Evidently, 10 μM 24 was fully replaced by 10 μM 83 demonstrating that both fragments targeted the same druggable pocket in BambL and showed a potential of improved binding over 24 (Figure 3 d). Interestingly, six closely related analogues of 83 (99–104, Table S2) confirmed the importance of the tetrazole (100) and morpholine (104) groups for binding of 83 to the secondary site given the lack of binding in TROSY NMR (Figure S14 b). Thus, 83 will be investigated in the future studies.
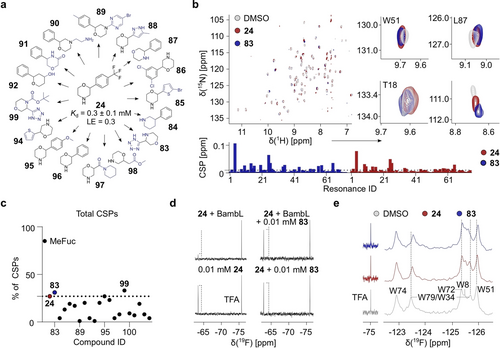
Structure–activity relationship study of 24. (a) Shown are 16 out of 22 commercial structural derivatives of 24, which were ranked using TROSY NMR. (b) TROSY NMR (top panel) of 15N BambL in presence of DMSO (gray), 83 (blue) or 24 (red). Qualitative analysis of TROSY NMR (bottom panel). Dashed line indicates CSPs>0.01 ppm. (c) Total % of CSPs derived in TROSY NMR shows that 83 (31 %) and 99 (33 %) promoted more CSPs in 15N BambL compared to initial hit 24 (dashed line, 27 %). (d) 0.1 mM 83 displaced 0.1 mM 24 from its binding site in 19F CPMG NMR. (e) PrOF NMR of 0.1 mM 5FW BambL with 1 mM 83 and 1 mM 24, which showed CSPs of 5FW resonances (dashed line) demonstrating the effect of both fragments on the orthosteric site of 5FW BambL.
Computational docking analysis of five derivatives of 24 (83, 84, 87, 90 and 94) was performed to check if the predicted pocket in BambL could accommodate these compounds. Hereby, we chose these fragments to check the importance of the morpholine and benzyl groups. As result, all compounds could be accommodated in the site, whereas 84 did not form electrostatic interactions (Figure S15). These observations were in agreement with our experimental data showing the role of the morpholine group (24) in BambL binding as shown for 87, whereas other parts of 24 are rather interchangeable (e.g. 83 and 94). Next, we confirmed that the analogue 83 could affect the orthosteric site of 5FW BambL in PrOF NMR (Figure 3 e). Indeed, 83 perturbed W79/W34 (W2 and W5), W51 and W72 similar to the initial hit 24 causing even larger NMR chemical shift perturbations of 5FW resonances.
Taken together, our SAR study confirmed the presence of a druggable secondary site in BambL. Moreover, binding of two fragments with a similar scaffold (24 and 83) to the secondary pocket of BambL affected its orthosteric site suggesting a communication between both sites.
Communication between the Carbohydrate and Remote Sites in BambL
To further prove allosteric communication in BambL, we proposed that mutations in the orthosteric and secondary sites could introduce perturbations that propagate through the network and result in similar chemical shift changes. For this, we used four (W8F, W51F, W72F and W74F) and three (T18S, T25S and L87R) mutants for the orthosteric and predicted remote sites, respectively (Figure 4 a). All mutants were folded and active as observed by TROSY NMR (Figure S16). Next, we quantified and compared the NMR chemical shift perturbations (CSPs) induced by mutations in their apo forms with respect to the wild type (WT). Interestingly, perturbations were not restricted to residues in the close periphery, but also affected remote residues, as shown for W51F and T18S mutants. Here, we clearly identified a chemical shift perturbation of W72 in both WT and T18S mutants moving along the same vector (Figure 4 b). Quantification of NMR chemical shift perturbations of the apo WT to other apo mutant forms (W8F, W72F, W74F, L87R and T18S) revealed the conformational changes through the same paths in 15N BambL, which is typical for allosteric proteins (Figure 4 c).27 Further, we assessed whether mutations in the predicted pocket alter the affinity of proteins to the natural carbohydrates, we titrated 2FF and a complex carbohydrate (F–H type 2) to BambL WT and T18S (Figures 4 d and S17 a). Compared to BambL WT, T18S preserved its affinity for 2FF (Figure 4 e, WT: Kd=7.9±0.2 μM, T18S: Kd=8.2±0.2 μM). However, BambL T18S showed nearly two-fold decrease in affinity for a complex carbohydrate, F–H type 2 (Figure S17 b, Kd=16.7±2.5 μM) compared to BambL WT (Kd=9±2 μM28). To verify the binding epitope of F–H type 2 to 15N BambL T18S, we used TROSY NMR and compared it to WT (Figure S17 c). Overall, T18S mutation reduced the magnitude of CSPs in 15N BambL suggesting a negative modulatory role of the pocket on the orthosteric site in recognizing complex carbohydrates (Figure S17 d). Interestingly, a discrepancy between two carbohydrate-binding sites in the interaction with the complex carbohydrates, but not with MeFuc, has been reported for BambL recently.9a Given the lack in affinity change with 2FF, we propose that inhibition of secondary site could potentially down-regulate the affinity of BambL by tuning the orthosteric site between two monomers, but not within a monomer. However, the hexameric structure of BambL complicates the identification of allosteric pathway since the symmetry-related sites are close to each other and chemical shifts can be affected by two binding events. As reported previously for RSL,29 it could be envisaged in the future to engineer a single monomeric neo-BambL with controlled number and position of binding sites in order to separate the effects on different sites. Thus, this hypothesis requires further investigations.
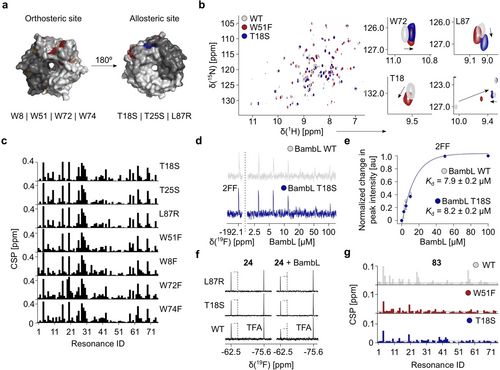
Characterization of the secondary site in BambL. (a) Top and bottom views on the orthosteric (red) and potential allosteric site (blue) in the crystal structure of BambL in complex with l-fucose (orange, PDB ID: 3ZZV). Single-point mutations in the orthosteric and secondary site have been proposed to assess the communication between the two sites. (b) Overlay of 15N TROSY NMR spectra of BambL WT, W51F and T18S shows the conformational changes introduced by both site-directed mutations. Notably, W51F and T18S mutations promoted identical changes on other resonances in the orthosteric (W72) and secondary sites (L87R) in BambL. (c) CSP studies of mutant apo forms compared to BambL WT show a preserved CSP pattern in both orthosteric and allosteric pocket mutants. (d) 19F NMR spectra of 2FF in presence of BambL WT and T18S. (e) Determination of 2FF Kd values for BambL T18S revealed a preserved affinity compared to BambL WT. (f) 19F CPMG NMR of 24 with BambL WT, T18S and L87R verified the predicted site given the lack of change in the peak intensity in mutants (dashed line). (g) Binding of 83 to W51F and T18S promoted less total CSPs above the threshold of 0.01 ppm compared to WT supporting the existence of a communication between the orthosteric and the remote sites.
Finally, we examined the impact of the pocket mutations on binding of the most potent fragments 24 and 83 by 19F CPMG and TROSY NMR. Notably, pocket mutations reduced 24 binding in 19F CPMG (Figure 4 f) given the lack of the peak intensity reduction in BambL T18S and L87R. Moreover, 24 promoted less CSPs above the threshold of 0.01 ppm in both mutants in TROSY NMR experiments (Figure S18 a,b) allowing us to conclude that the mutations blocked the entrance into the predicted pocket only partially. Similarly, we observed this effect with the fragment 83 (Figure S19 a), which is in agreement with the computational docking analysis suggesting the presence of two orientations for 83 and its derivative 99 (Figure S20). Interestingly, mutation in the orthosteric site (W51F) reduced 83 binding similarly to 15N BambL T18S and other pocket mutants (Figures 4 g and S19 b), which confirms the “end-to-end” communication of both sites.
Taken together, these data reveal the existence of a druggable allosteric site in BambL. The NMR chemical shift perturbations of backbone resonances of 15N BambL pocket mutants are located in sites distal to the actual pocket and the communication extends to the carbohydrate recognition site, suggesting a propagation of conformational changes in BambL upon changes in the allosteric pocket.
Conclusion
We report the presence of druggable pockets in a bacterial β-propeller lectin BambL, which could be used to design allosteric inhibitors. We showed binding of fragments to BambL in a 19F NMR screening and validated hits using orthogonal methods: SPR and TROSY NMR. Computational pocket prediction analysis SiteMap identified three potential druggable pockets in BambL trimer. We also showed that the potential secondary binding sites could accommodate drug-like molecules (24, 10 and 12). Initial SAR study of 24 (Kd=0.3±0.1 mM, LE=0.3) proved the pocket identity by confirming the predicted part of 24 scaffold responsible for its binding to the pocket in our docking study. Notably, fragment binding to the secondary site induced conformational changes in the orthosteric site of 5FW BambL in PrOF NMR. This observation allowed us to propose the presence of a communication between two spatially distant binding sites in BambL. Employing site-directed mutagenesis within the predicted and orthosteric sites, we observed conformational changes of 15N BambL backbone resonances in TROSY NMR in distal regions from the mutation sites. Such behavior is typical for allosteric proteins. Given a fungal AFL and bacterial RSL lectins show similarities to BambL in structure, hit rates and secondary pockets, we believe the allostery could also be present in other β-propeller lectins. These observations will support future drug-discovery campaigns that aim to develop drug-like allosteric inhibitors for bacterial and fungal lectins.
Acknowledgements
We thank the Max Planck Society and German Research Foundation (DFG) [RA1944/7-1] for supporting this work, which was in the scope of German Research Foundation and French National Research Agency [ANR-17-CE11-0048] project “Glycomime”. The authors acknowledge support by the H2020 PhD4Glycodrug program (MSCA 76558), ANR PIA Glyco@Alps (ANR-15-IDEX-02) and Labex Arcane/CBH-EUR-GS (ANR-17-EURE-0003).
Conflict of interest
The authors declare no conflict of interest.