Innovation by Evolution: Bringing New Chemistry to Life (Nobel Lecture)†
Copyright © The Nobel Foundation 2018. We thank the Nobel Foundation, Stockholm, for permission to print this lecture.
Graphical Abstract
The directed evolution of enzymes is now routinely used to develop new catalysts with various applications, such as in environmentally friendly production of chemicals and renewable fuels. In her Nobel lecture, F. Arnold describes how lessons from nature inspired the development of methods for directed evolution.
I am a chemist and engineer who looks upon the living world with the deepest admiration. Nature, herself a brilliant chemist and by far the best engineer of all time, invented life that has flourished for billions of years under an astonishing range of conditions. I am among the many inspired by the beauty and remarkable capabilities of living systems, the breathtaking range of chemical transformations they have invented, the complexity and myriad roles of the products. I am in awe of the exquisite specificity and efficiency with which Nature assembles these products from simple, abundant, and renewable starting materials. Where does this chemistry come from? It derives from enzymes, the DNA-encoded protein catalysts that make life possible, molecular machines that perform chemistry no human has matched or mastered.
Evolution, a Grand Diversity-Generating Machine
Equally awe-inspiring is the process by which Nature created these enzyme catalysts and in fact everything else in the biological world. The process is evolution, the grand diversity-generating machine that created all life on earth, starting more than three billion years ago. Responsible for adaptation, optimization, and innovation in the living world, evolution executes a simple algorithm of diversification and natural selection, an algorithm that works at all levels of complexity from single protein molecules to whole ecosystems. No comparably powerful design process exists in the world of human engineering.
I wanted to engineer Nature's enzymes to make ones tailored to, and uniquely suited for, human purposes. For close to five thousand years we have made use of microbial enzymes to brew beer and leaven bread. Once the protein catalysts were identified and isolated, many more diverse applications were devised. Today, enzymes are used to diagnose and treat disease, reduce farm waste, enhance textiles and other materials, synthesize industrial and pharmaceutical chemicals, and empower our laundry detergents. But so much more could be achieved if we understood how to build new ones.
Early protein engineers struggled mightily with this goal. In those days (the 1980s), we did not know enough about how a DNA sequence encodes enzyme function to design enzymes for human applications. Unfortunately, this is still true: today we can for all practical purposes read, write, and edit any sequence of DNA, but we cannot compose it. The code of life is a symphony, guiding intricate and beautiful parts performed by an untold number of players and instruments. Maybe we can cut and paste pieces from nature's compositions, but we do not know how to write the bars for a single enzymic passage. However, evolution does.
Exploring the Universe of Possible Proteins
Some researchers think of the protein universe as the set of all proteins that Nature has devised. But these proteins, relevant to biology, are an infinitesimal fraction of the possible proteins. The universe of possible proteins, my universe, contains solutions to many of humanity's greatest needs: there we will find cures for disease, solutions to energy crises and a warming world, food and clean water for a growing population, and ways to arrest the miseries of aging. I wanted to explore this universe to find those proteins that will serve humanity.
But how does one discover a useful protein in the infinitude of possible proteins, a set larger by many orders of magnitude than all the particles in the universe? In his fascinating short story, the Library of Babel, Jorge Luis Borges describes a collection comprising all possible books assembled from an alphabet of letters.1 Most texts in Borges’ library are gibberish, and his despairing librarians, for all their lifelong efforts, cannot locate a single meaningful sentence, much less a complete story.
Similarly, most possible protein sequences encode nothing we would recognize as meaningful. Unlike Borges’ librarians, however, I am entirely surrounded by proteins with meaningful stories. They are everywhere and can literally be scraped from the bottom of my shoe, captured from the air I breathe, or extracted from a database. These are the products of billions of years of work performed by mutation and natural selection. And evolution continues to create new ones from these rare functional sequences that were themselves discovered by evolution. Thus I decided to start my exploration by using this gift from evolution, the existing functional proteins.
There are thousands of ways to make one change in the amino acid sequence of a protein. There are millions of ways to modify it by two changes, and so on—the numbers grow so rapidly that making a single copy of each protein altered by only 1 % of its sequence would require the weight of the world in materials. And the vast majority of these modified sequences are neither usable nor useful. The challenge therefore is to discover protein sequences that provide new benefits and deliver novel improvements on a thrifty scale of weeks, rather than millennia or eons, and with the help of one graduate student rather than that of an army. To outperform Nature, I needed a strategy that sidesteps the despair of the Babel librarians.
John Maynard Smith helped answer this challenge for me in a beautiful paper published in 1970.2 Consider an ordered space in which any protein sequence is surrounded by neighbors that have a single mutation. For evolution to work, he reasoned, there must exist functional proteins adjacent to one another in this space. Although most sequences do not encode functional proteins, evolution will work even if just a few meaningful proteins lie nearby. Given low levels of random mutation, the filter of natural selection can find those sequences that retain function. In fact, many of today's proteins are the products of a few billion years of mostly such gradual change. Many of these mutations are neutral and change little, but others can be deleterious. Natural selection picks the wheat from the chaff and guides mutating proteins along continuously functional paths through the vast space of sequences mostly devoid of function.
But by using evolution I want to make better proteins, proteins that serve my purposes. Thus directed protein evolution becomes a search on a new fitness landscape, where fitness is performance and is defined by the artificial selection I impose. This is a landscape whose structure we knew very little about in the 1980s. Evolution on a rugged landscape is difficult, as mutation propels sequences into crevasses of non-function. However, latching onto Maynard Smith's argument that proteins evolve on a landscape smooth in at least some of its many dimensions, I reasoned that directed evolution could find and follow continuous paths leading to higher fitness.3
A Process for Evolving Proteins in the Laboratory
Science, like all human endeavors, is evolutionary. We progress by adding to and recombining what is present. Important developments in the 1980s and 1990s influenced my thinking. Manfred Eigen speculated on in vitro molecular evolution,4 and Gerry Joyce was selecting RNA “enzymes”’ that could cleave DNA from pools of billions, perhaps trillions, of mutated sequences.5 Error-prone PCR (polymerase chain reaction)5, 6 became a useful tool for random mutagenesis of genes. Jim Wells7 and others demonstrated that beneficial mutations in proteins could be accumulated. Stuart Kauffman quantified evolutionary trajectories on model fitness landscapes,8 and the philosopher Daniel Dennett supplied the conceptual framework that helped me convey the power of evolution to others.9
Protein genotypes and phenotypes do not coexist in one molecule, as they do for RNA, and protein fitness landscapes differ fundamentally from those of RNA. Thus directed protein evolution would require different strategies and experimental tools. To devise a directed evolution strategy suitable for enzymes, I started with the fundamental rule: “You get what you screen for.”
We were generating enzymes of interest in recombinant microorganisms by inserting genetic material that we could mutate in the test tube. We used common microbes like Escherichia coli or yeast to produce “libraries” of mutant enzymes to test for desired functions. Since we were making enzymes for human applications, we rejected microbial growth or survival selections favored by microbiologists and geneticists. While those approaches enable a straightforward search through thousands and even millions of variants in one experiment, they do not meet our criteria of affording function in novel environments, over-expression in a production host, compatibility with new substrates, specific product formation, and so on. Thus we turned to good old-fashioned analytical chemistry to develop reproducible, reliable screens that reported what mattered to us.
To measure what mattered, we were limited to monitoring the few thousand protein variants we could express and array in readily available 96-well plates or on a petri dish. Therefore we could only search deeply those sequences one or two mutations away from the starting protein. Given that such a small change in sequence would be expected to generate only small improvements in function, we would have to deploy reproducible screening assays capable of finding those rare and only slightly improved protein progeny. A desirable mutation might yield only a two-fold increase in catalytic activity or a few degrees’ step up in melting temperature. To achieve significant changes, we would have to multiply those benefits over successive generations.
This strategy works well when re-optimizing enzymes for new tasks. While a natural enzyme generally performs well in its biological job, it is often less enthusiastic about doing a new job and initially works poorly (Figure 1). New demands change the fitness landscape, often knocking a protein down from a position that was painstakingly acquired through the work of natural evolution. Sequential rounds of random mutation and screening for improved performance, however, can accumulate the beneficial mutations needed to climb to a new peak.
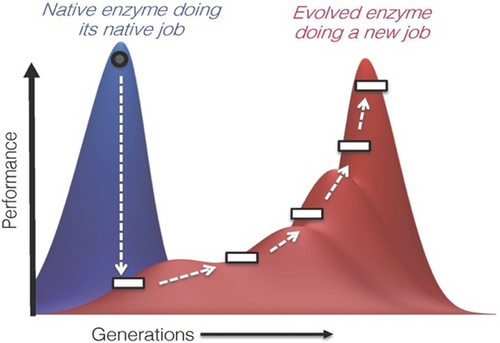
An enzyme whose function is optimized for its native job generally performs poorly in a new role. Directed evolution through rounds of mutation and screening can discover changes in sequence that improve performance, climbing a new fitness peak.
To illustrate, in the late 1980s my research group started to re-engineer a protease, subtilisin E, to perform its hydrolytic reaction under unusual and non-natural conditions. We chose to have the enzyme function in high concentrations of a polar organic solvent (dimethylformamide, DMF) that causes wild-type subtilisin E to lose most of its activity. We used random mutagenesis and screening to recover activity lost by adding low concentrations of DMF, combining the beneficial mutations.10 Emboldened by these results, Keqin Chen performed iterations of random mutagenesis and screening for activity in increasing concentrations of the organic solvent and evolved an enzyme that performed as well in 60 % DMF as its wild-type parent did in the absence of DMF, a 256-fold increase in activity.11
Strikingly, this enzyme adapted rapidly to a challenge it presumably had not encountered during its evolution. Furthermore, the mutations that led to the improved performance were unexpected. We could not explain how mutations located on loops surrounding the enzyme's active site enhanced activity in high concentrations of organic solvent, much less plan them in a rational approach to engineering an enzyme with this new capability. But we had a process that gave the right result, even if that result would require much more reverse engineering to understand fully.
I met Pim Stemmer at a workshop organized by Stuart Kauffman at the Santa Fe Institute in 1995, not long after his landmark “DNA shuffling” paper was published.12 Pim introduced sex—recombination—as a search strategy for protein evolution and called his method molecular breeding, a description I now often use to explain what I do. At Maxygen, the company he started in 1997 that licensed our technologies, and where I served on the founding Science Advisory Board, Pim's vision was grand: he wanted to evolve viruses, metabolic pathways, plant traits, and human therapeutics. My focus was entirely on enzymes and getting useful results quickly.
Those results ensued. A few examples of how enzymes could be evolved to accept challenging, non-natural substrates13 or function at high temperatures,14, 15 work of intrepid lab members Jeffrey Moore, Huimin Zhao, and Lori Giver convinced many researchers, especially those in industry where deadlines were tight and interest in understanding why individual mutations were beneficial lagged behind the need for the enzyme. Directed evolution offered a reliable optimization algorithm: find a starting enzyme, develop a moderate-throughput assay, and turn the crank.
The methods we developed and demonstrated in the 1990s were adopted rapidly. That decade saw the explosive rise of directed evolution in industrial and academic laboratories around the world, especially those of Andy Ellington, Manfred Reetz, Uwe Bornscheuer, George Georgiou, Romas Kazlauskas, and Don Hilvert, who introduced many novel concepts and improvements. In its original and in many modified forms, directed evolution produces new gene editing tools, therapeutic enzymes, and enzymes for diagnostics, DNA sequencing and synthesis, imaging, agriculture, textiles, cleaning aids, and much more. Some of those developments are detailed in excellent reviews (e.g., refs. 16, 17).
What important lessons did we learn about enzymes from the early directed evolution experiments? First and foremost, we learned that enzymes can adapt to new challenges. It often takes only a few mutations for an enzyme to acquire the targeted trait. We had not known when we started out how many generations would be needed to obtain useful changes in function. Nature, after all, takes a circuitous route to achieving new properties, combining neutral or even negative mutations with beneficial ones. Those paths can involve hundreds of changes. Our approach, however, collected only adaptive mutations that yield steep changes in function. Useful traits could emerge in less than ten, or even five, generations.
We also learned that much remains to be done before we can reliably design good enzymes. Beneficial mutations found by directed evolution are often far from the site of catalysis. Even today we struggle to explain their effects, and are unable to predict them reliably or easily. Nevertheless, practitioners now enjoy a dependable process for improving enzymes that does not require us to understand their structures, folding, or catalytic mechanisms.
Evolution of Enzymes for Reactions Invented by Chemists
What fascinates me today is the evolution of new enzymes. I wish to go beyond optimizing biological functions that are already known, and instead bring to life whole new chemistries. But how can one create enzymes that catalyze reactions invented by chemists? One cannot go to a biochemical data base to find enzyme sequences annotated for such transformations. For many years, in fact, creating new chemistry by directed evolution seemed to me an insurmountable challenge. Enzymes position functional groups in exquisite arrangements to bind substrates and stabilize reaction transition states. For a long time I could not see how my conservative directed evolution strategy of accumulating one or two beneficial mutations per generation would create entirely new enzyme active sites. Unless, of course, an active site is already largely there…
When innovating, Nature does not invent new active sites de novo. Rather, to support the fight for survival or to move into a new niche, emerging enzymes exploit existing catalytic mechanisms and machineries.18 The biological world is replete with proteins whose chemical capabilities extend well beyond the functions for which they are selected at any given time. These “promiscuous” activities can become advantageous, such as when a new food source becomes available, and provide the basis for evolution of a new enzyme that gives a fitness advantage to its host.19 Promiscuous functions can also be useful for human applications.20 If a new catalytic activity is already present, even at a low level, our conservative process of accumulating beneficial mutations can mold it into a new enzyme. Dan Tawfik, in particular, compellingly demonstrated how known promiscuous enzyme activities, sometimes relics of their own ancestral origins, may be evolvable in the laboratory.21 Directed evolution can innovate when the innovation is already present. For several years we have capitalized on this realization to create whole families of enzymes that catalyze reactions previously unknown in biology.22
To explain this process, we turn to the cytochrome P450 enzyme family. Nature draws on the P450′s highly reactive iron-oxo Compound I (and other intermediates) to perform varied reactions that presumably evolved from the promiscuous functions of ancestral P450s (Figure 2). Today, the cytochrome P450 family has members that can transfer an oxygen atom to organic molecules to make specific hydroxylated compounds or epoxides, oxidize heteroatoms, nitrate aromatics, and much more. The biological world shaped these enzymes using the diversity-generating machine of evolution, and hundreds of thousands of their sequences are stored in databases.
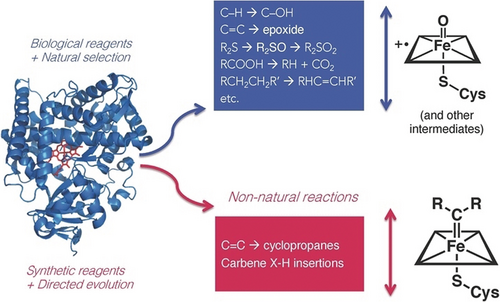
Expanding the scope of P450 chemistry. The cytochrome P450 family, whose members were presumably created by gene duplication and natural selection of promiscuous functions, comprises enzymes that use reactive oxygen intermediates to catalyze a wide range of reactions. We reasoned that we could expand the scope of P450 chemistry by using synthetic carbene and nitrene precursors to drive formation of new reactive intermediates. Directed evolution would be used to mold the enzyme, controlling and enhancing new-to-nature activities.
This magnificent biological diversity now drives laboratory innovations. With insights and inspiration from chemistry, directed evolution can take us where biology has never gone. For instance, if a P450 can transfer reactive oxygen species to substrates, perhaps it can also be directed to transfer reactive nitrogen or carbon species and assemble molecules using efficient strategies hitherto unused by biology. To Pedro Coelho and Eric Brustad, members of my lab in 2012, the P450 enzyme's reactive Compound I intermediate resembled an iron carbenoid, long used by chemists to transfer carbenes to carbon–carbon double bonds in alkenes, or participate in X−H insertion reactions to form new heteroatom-carbon bonds (Figure 2). These reactions, unknown in biology, are possible in chemistry when humans supply synthetic diazo carbene precursors and transition metal catalysts modeled after heme cofactors.
We thought perhaps by offering man-made carbene precursors to heme proteins we could discover promiscuous “carbene transferase” activities.23 If so, we might use directed evolution to draw out and improve such biologically irrelevant but synthetically interesting capabilities.
An early achievement for this approach was alkene cyclopropanation, a transformation well known in transition metal catalysis but unknown in biology. Inspired by early reports of heme mimics catalyzing carbene transfer to alkenes in organic solvents, we discovered that iron-heme proteins do indeed promote cyclopropanation when provided with diazo carbene precursors and a suitable alkene substrate, in water. Furthermore, mutations altered both the activity and the selectivity of product formation so that enzymes produced individual cyclopropane stereoisomers.23
This new reaction has many practical applications. For example, directed evolution resulted in a highly efficient enzyme for production of the chiral cis-cyclopropane precursor to the antidepressant medication levomilnacipran.24 We and Rudi Fasan have since engineered a variety of heme proteins to synthesize other pharmaceutical precursors.25, 26 Because alkene cyclopropanation proceeds in whole Escherichia coli cells that express the evolved enzyme, as well as in cell lysate, preparing the catalyst is as simple as growing bacteria.
At the same time, we also discovered that some engineered cytochromes P450 performed nitrene chemistry, generating the iron-nitrenoid from a synthetic azide nitrene precursor and directing the nitrene to C−H bonds for C−H amination (Figure 2).27 Earlier chemical research stimulated these experiments as well. In 1985, Gellman and co-workers reported that supplying an iminoiodane nitrene precursor to a rabbit liver cytochrome P450 led to three turnovers of intramolecular C−H amination.28 We and Rudi Fasan re-discovered this promiscuous nitrene transfer activity at more or less the same time, almost thirty years later.27, 29 More enzymes catalyzing abiological nitrene transfer reactions followed, brought about by a combination of chemical insight for reaction discovery and directed evolution to improve nascent activities.30
Since a goal of this lecture is to introduce foundational concepts and explain how we came to them, rather than to review research results, I will mention just one recent example of making products that chemists find very challenging: highly strained rings. Producing bicyclobutanes by two carbene transfers to an alkyne is a transformation not known in biology. It is rare in the world of human chemistry, and was never reported to be catalyzed using iron. Kai Chen first evolved an engineered, serine-ligated cytochrome P450 to transfer a carbene to an alkyne with perfect selectivity and make single stereoisomers of cyclopropenes. These carbocycles, whose ring strain is greater than 50 kcal mol−1, are highly challenging to synthesize stereoselectively, but the enzyme does it with ease. Using the appropriate alkyne, Kai Chen also coaxed the enzyme to transfer a second carbene, cyclopropanating the double bond of the cyclopropene in the protected enzyme active site to make bicyclobutanes having >60 kcal mol−1 of ring strain (Figure 3). Following directed evolution, the enzymes delivered single stereoisomers of highly strained cyclopropenes or bicyclobutanes with turnovers in the thousands.31
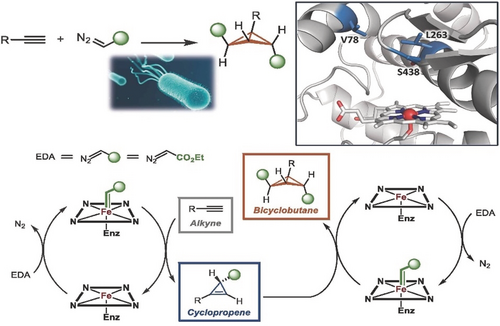
Enzyme-catalyzed bicyclobutanation through two carbene transfers to an alkyne, catalyzed by a serine-ligated variant of a cytochrome P450.31 The enzyme must generate the reactive carbene and transfer it to the alkyne substrate and then do it a second time to the cyclopropene intermediate in order to generate the bicyclobutane. Evolved enzymes make a single stereoisomer of these highly strained rings, which indicates a well-defined orientation of the substrates in the newly-evolved active site.
When supplied with alkynes and carbene precursors, E. coli expressing these new enzymes churn out cyclopropenes and bicyclobutanes. For sugar and a few growth-promoting trace elements, these living catalysts perform their chemistry in water (buffer), at room temperature. We hope their remarkable selectivities, low cost, and ability to use Earth-abundant iron to make strained rings that are otherwise difficult to obtain will open new applications for these fascinating products.
Bringing New Bonds to Biology
For a final glimpse into the exciting future chemistry that laboratory-evolved enzymes will enable, I will describe how we can now create biocatalytic machinery to make bonds unknown in biology. Silicon is the second most abundant element in the Earth's crust. Despite their ubiquity, carbon-silicon bonds are non-existent in the biological world. Yet laboratories make them, and lots of them. The room in which you are reading contains caulks, sealants, earphones, hair gels, and many more products whose carbon–silicon bonds are human-made.
In 2016, Jennifer Kan and her team discovered that heme proteins can catalyze carbene insertion into Si−H bonds to make various organosilicon products.32 We are particularly fond of a marvelous little cytochrome c from Rhodothermus marinus, isolated from a hot saltwater pool in Iceland and now in the Protein Data Bank. This cytochrome c holds onto its heme through a covalent attachment; it is a manageable 124 amino acids long, its three-dimensional structure is known, and it is hyper-stable. With a melting temperature above 100 °C, it can even be boiled and not lose its fold or metal cofactor. Although its biological function is electron transfer, it also happens to catalyze Si−H insertion: Rma cyt c inserts the carbene derived from methyl ethyldiazoacetate into dimethylphenylsilane with 40 turnovers and 97 % enantiomeric excess (ee).
Those who know cytochromes c may find this puzzling. The iron in a cytochrome c is coordinatively saturated, i.e., four equatorial nitrogen ligands come from the porphyrin ring, while in Rma cyt c a methionine and a histidine provide the two axial ligands. Hence it is reasonable to ask where the reactive carbene forms, and how this protein binds the silane substrate. In fact, measuring the volume of the active site by rolling a computer ball over the crystal structure delivers an answer of zero.
Yet nature cares nothing for our calculations. This protein catalyzes its Si−C bond-forming reaction almost as effectively as the best human-invented catalysts for a similar reaction (which, by the way, use precious metals rather than readily-available iron), and it evolves. Just three generations of mutations directed to residues in the active site and screening for higher activity generated an enzyme now 15 times more active than human-invented catalysts. The new enzyme makes single enantiomers of its products and has a good substrate scope, producing new organosilicon compounds from at least 20 different silanes with hundreds to thousands of turnovers and >99 % ee.32 Once again, the enzyme is fully genetically encoded, so that bacteria expressing the gene form new silicon–carbon bonds, perhaps for the first time ever in a living system. Of course, we cannot know for sure that it is the first time, given that so much chemistry of the biological world remains unexplored.
A wonderful feature of engineering by evolution is that solutions come first; an understanding of the solutions may or may not come later. The evolved protein can be studied biochemically in an effort to discern how its new features emerged. In the case of the Si−H insertion enzyme derived from Rma cytochrome c, the X-ray crystal structure showed that the three activity-enhancing mutations changed the structure of a loop over the iron. What was the methionine axial ligand in the wild-type protein became an aspartic acid that no longer ligated the iron and instead pointed out to the solvent. A flip in the configuration of the loop created a binding pocket and also made the loop more dynamic so that it accommodates both the carbene and a range of silane substrates. Rusty Lewis was even able to trap the reactive carbene in the evolved enzyme and observe its orientation in the protein crystal at high resolution. Structural and spectroscopic studies combined with computational models allowed us to begin to explain how the new catalytic activity arose.33 That does not mean, however, that we could predict the mutations that produced these effects.
Why stop at silicon? Another element of interest is boron, richly represented in the deserts not far from my southern California home. As with silicon, a wide range of organoboron compounds have been created in laboratories, but carbon-boron bonds have never been found in the biological world. Biology uses boron in the form of borates incorporated into natural products, most likely without the help of specific enzymes. Similarly, silicates are widely found in plants and marine animals such as diatoms.
To make carbon–boron bonds biologically, we again turned to our favorite cytochrome c. Jennifer Kan and Xiongyi Huang uncovered some activity for forming organoboron compounds by carbene insertion in the B−H bonds of water-stable borane adducts. Directed evolution created an enzyme that catalyzed 400 times more turnovers than the best small-molecule catalysts reported for similar transformations. Again, the enzyme is fully genetically encoded and carries out this new function inside living bacteria.34
Carbon–X bonds known in biology include mostly C−H, C−N, C−O, C−S bonds, some bonds to halogens, and a few to P, As, Se, and some metals (Figure 4). This leaves vast swaths of the periodic table untouched. With help from directed evolution and inspiration from the transition metal catalysis literature, two whole new elements have been added to biology's C−X bond repertoire. This ability can be exploited to bring boron and silicon into life and into products that can be manufactured in engineered microorganisms. The enzymes can also be used in synthetic chemistry, where their exquisite and tunable selectivities will enable low-cost and easy preparation of organoboron and organosilicon products.
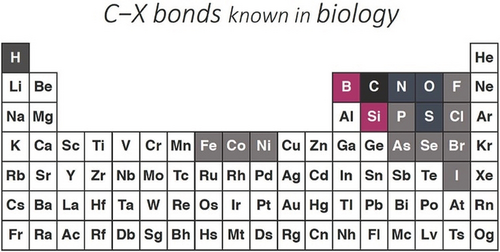
New “carbene transferases” made by directed evolution have added C−Si and C−B bonds to biology's DNA-encoded synthetic repertoire.
Final Thoughts About the Future of Directed Evolution
Life—the biological world—is the greatest chemist, and evolution is her design process. In fact, the internet of living things has been crowd-sourcing problem solving this way for more than three billion years. Evolution can circumvent our profound ignorance of how sequence encodes function, and may allow us to find new solutions to human problems. I have described how we are now moving into a future of readily-tunable catalysts that perform challenging reactions using Earth-abundant materials and producing minimal waste. Biological systems are good models for sustainable chemistry that uses abundant, renewable resources and recycles a good fraction of its products. I dream of the day that much of our chemistry becomes genetically encodable, and microorganisms and plants are our programmable factories.
I am continually amazed at the ease with which evolution innovates. With the power of evolution realized for engineering, we can look at diverse products of natural evolution in an entirely new way. Instead of asking what enzymes do in the natural world, we can now ask, “What might they do?” Enzymes will perform chemistry in more ways than we could have imagined, especially when we use evolution to unleash their latent potential. A treasure trove of new enzymes awaits discovery for carrying out chemistry that we could not even contemplate just a few years ago.
Existing diversity provides the fuel for these innovations; both natural and directed evolution uses this diversity to solve challenges, exploit opportunities, and evade catastrophe. As countless examples from the natural world attest, the alternative to diversity is extinction.
Acknowledgements
I thank the Royal Swedish Academy of Sciences, my family, friends, colleagues, Sabine Brinkmann-Chen, Cheryl Nakashima, and especially my current and former students. I am grateful to the California Institute of Technology, where I was inspired to explore the unknown for new understanding and found the team with which to do it.
Biographical Information
Frances Arnold is the Linus Pauling Professor of Chemical Engineering, Bioengineering and Biochemistry at the California Institute of Technology, where she has been since 1986. She pioneered directed enzyme evolution and has used those methods to create enzymes for applications in alternative energy, chemicals, and medicine. She received her B.S. in Mechanical and Aerospace Engineering from Princeton University and worked at the Solar Energy Research Institute, a national laboratory devoted to alternative energy research (now NREL) before obtaining her Ph.D. in Chemical Engineering from the University of California, Berkeley. She carried out postdoctoral studies at UC Berkeley and Caltech before her appointment as Assistant Professor of Chemical Engineering in 1987.