Amphipathic DNA Origami Nanoparticles to Scaffold and Deform Lipid Membrane Vesicles†
Dr. Aleksander Czogalla
Biotechnology Center of the TU Dresden, Tatzberg 47/51, 01307 Dresden (Germany)
Department of Cytobiochemistry, Faculty of Biotechnology, University of Wrocław ul. F. Joliot-Curie 14a, 50383 Wrocław (Poland)
Search for more papers by this authorDominik J. Kauert
Institute for Molecular Cell Biology, University of Münster, Schlossplatz 5, 48149 Münster (Germany)
Search for more papers by this authorDr. Henri G. Franquelim
Department of Cellular and Molecular Biophysics, Max Planck Institute of Biochemistry, Am Klopferspitz 18, 82152 Martinsried (Germany) http://www.biochem.mpg.de/en/rd/schwille/
Search for more papers by this authorDr. Veselina Uzunova
B CUBE—Center for Molecular Bioengineering, Dresden, University of Technology, Arnoldstraße 18, 01307 Dresden (Germany)
Search for more papers by this authorDr. Yixin Zhang
B CUBE—Center for Molecular Bioengineering, Dresden, University of Technology, Arnoldstraße 18, 01307 Dresden (Germany)
Search for more papers by this authorProf. Ralf Seidel
Institute for Molecular Cell Biology, University of Münster, Schlossplatz 5, 48149 Münster (Germany)
Search for more papers by this authorCorresponding Author
Prof. Petra Schwille
Department of Cellular and Molecular Biophysics, Max Planck Institute of Biochemistry, Am Klopferspitz 18, 82152 Martinsried (Germany) http://www.biochem.mpg.de/en/rd/schwille/
Department of Cellular and Molecular Biophysics, Max Planck Institute of Biochemistry, Am Klopferspitz 18, 82152 Martinsried (Germany) http://www.biochem.mpg.de/en/rd/schwille/Search for more papers by this authorDr. Aleksander Czogalla
Biotechnology Center of the TU Dresden, Tatzberg 47/51, 01307 Dresden (Germany)
Department of Cytobiochemistry, Faculty of Biotechnology, University of Wrocław ul. F. Joliot-Curie 14a, 50383 Wrocław (Poland)
Search for more papers by this authorDominik J. Kauert
Institute for Molecular Cell Biology, University of Münster, Schlossplatz 5, 48149 Münster (Germany)
Search for more papers by this authorDr. Henri G. Franquelim
Department of Cellular and Molecular Biophysics, Max Planck Institute of Biochemistry, Am Klopferspitz 18, 82152 Martinsried (Germany) http://www.biochem.mpg.de/en/rd/schwille/
Search for more papers by this authorDr. Veselina Uzunova
B CUBE—Center for Molecular Bioengineering, Dresden, University of Technology, Arnoldstraße 18, 01307 Dresden (Germany)
Search for more papers by this authorDr. Yixin Zhang
B CUBE—Center for Molecular Bioengineering, Dresden, University of Technology, Arnoldstraße 18, 01307 Dresden (Germany)
Search for more papers by this authorProf. Ralf Seidel
Institute for Molecular Cell Biology, University of Münster, Schlossplatz 5, 48149 Münster (Germany)
Search for more papers by this authorCorresponding Author
Prof. Petra Schwille
Department of Cellular and Molecular Biophysics, Max Planck Institute of Biochemistry, Am Klopferspitz 18, 82152 Martinsried (Germany) http://www.biochem.mpg.de/en/rd/schwille/
Department of Cellular and Molecular Biophysics, Max Planck Institute of Biochemistry, Am Klopferspitz 18, 82152 Martinsried (Germany) http://www.biochem.mpg.de/en/rd/schwille/Search for more papers by this authorThis work has been supported in collaborative research centers of the Deutsche Forschungsgemeinschaft (DFG), begun in TRR83, and continued in SFB863. H.G.F. acknowledges the receipt of a Humboldt Research Fellowship. Further support was given by the Max Planck Society to P.S., the Leibniz Association and the BMBF (grant numbers SAW-2011-IPF-2/380032 and 03Z2E511) to V.U. and Y.Z. as well as by the European Research Council (grant number GA 261224) and by the DFG (TRR61) to R.S.
Graphical Abstract
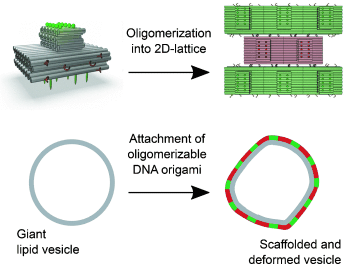
Amphipathic DNA origami structures were engineered, which have a flat membrane-binding interface decorated with cholesterol-derived anchors and sticky oligonucleotide overhangs enabling formation of ordered arrays on a membrane. Such DNA origami structures are capable of deforming free-standing lipid membranes (see picture), mimicking the biological activity of coat-forming proteins.
Abstract
We report a synthetic biology-inspired approach for the engineering of amphipathic DNA origami structures as membrane-scaffolding tools. The structures have a flat membrane-binding interface decorated with cholesterol-derived anchors. Sticky oligonucleotide overhangs on their side facets enable lateral interactions leading to the formation of ordered arrays on the membrane. Such a tight and regular arrangement makes our DNA origami capable of deforming free-standing lipid membranes, mimicking the biological activity of coat-forming proteins, for example, from the I-/F-BAR family.
Supporting Information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
| Filename | Description |
|---|---|
| anie_201501173_sm_miscellaneous_information.pdf1.2 MB | miscellaneous_information |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1
- 1aK. Farsad, P. De Camilli, Curr. Opin. Cell Biol. 2003, 15, 372–381;
- 1bM. M. Kozlov, F. Campelo, N. Liska, L. V. Chernomordik, S. J. Marrink, H. T. McMahon, Curr. Opin. Cell Biol. 2014, 29, 53–60.
- 2
- 2aA. Frost, V. M. Unger, P. De Camilli, Cell 2009, 137, 191–196;
- 2bO. Daumke, A. Roux, V. Haucke, Cell 2014, 156, 882–892.
- 3H. T. McMahon, J. L. Gallop, Nature 2005, 438, 590–596.
- 4B. Sorre, A. Callan-Jones, J. Manzi, B. Goud, J. Prost, P. Bassereau, A. Roux, Proc. Natl. Acad. Sci. USA 2012, 109, 173–178.
- 5A. Frost, R. Perera, A. Roux, K. Spasov, O. Destaing, E. H. Egelman, P. De Camilli, V. M. Unger, Cell 2008, 132, 807–817.
- 6
- 6aB. Qualmann, D. Koch, M. M. Kessels, EMBO J. 2011, 30, 3501–3515;
- 6bH. Zhao, A. Pykalainen, P. Lappalainen, Curr. Opin. Cell Biol. 2011, 23, 14–21;
- 6cA. Pykäläinen, M. Boczkowska, H. Zhao, J. Saarikangas, G. Rebowski, M. Jansen, J. Hakanen, E. V. Koskela, J. Peranen, H. Vihinen, E. Jokitalo, M. Salminen, E. Ikonen, R. Dominguez, P. Lappalainen, Nat. Struct. Mol. Biol. 2011, 18, 902–907.
- 7P. Schwille, Science 2011, 333, 1252–1254.
- 8
- 8aP. W. Rothemund, Nature 2006, 440, 297–302;
- 8bC. E. Castro, F. Kilchherr, D. N. Kim, E. L. Shiao, T. Wauer, P. Wortmann, M. Bathe, H. Dietz, Nat. Methods 2011, 8, 221–229.
- 9S. M. Douglas, H. Dietz, T. Liedl, B. Hogberg, F. Graf, W. M. Shih, Nature 2009, 459, 414–418.
- 10H. Dietz, S. M. Douglas, W. M. Shih, Science 2009, 325, 725–730.
- 11
- 11aK. Jahn, T. Torring, N. V. Voigt, R. S. Sorensen, A. L. Bank Kodal, E. S. Andersen, K. V. Gothelf, J. Kjems, Bioconjugate Chem. 2011, 22, 819–823;
- 11bF. C. Simmel, Curr. Opin. Biotechnol. 2012, 23, 516–521.
- 12M. Schade, D. Berti, D. Huster, A. Herrmann, A. Arbuzova, Adv. Colloid Interface Sci. 2014, 208, 235–251.
- 13
- 13aM. Langecker, V. Arnaut, T. G. Martin, J. List, S. Renner, M. Mayer, H. Dietz, F. C. Simmel, Science 2012, 338, 932–936;
- 13bS. D. Perrault, W. M. Shih, ACS Nano 2014, 8, 5132–5140;
- 13cY. Suzuki, M. Endo, Y. Yang, H. Sugiyama, J. Am. Chem. Soc. 2014, 136, 1714–1717;
- 13dJ. R. Burns, K. Gopfrich, J. W. Wood, V. V. Thacker, E. Stulz, U. F. Keyser, S. Howorka, Angew. Chem. Int. Ed. 2013, 52, 12069–12072; Angew. Chem. 2013, 125, 12291–12294.
- 14A. Czogalla, E. P. Petrov, D. J. Kauert, V. Uzunova, Y. Zhang, R. Seidel, P. Schwille, Faraday Discuss. 2013, 161, 31–43; discussion 113–150.
- 15A. Czogalla, D. J. Kauert, R. Seidel, P. Schwille, E. P. Petrov, Nano Lett. 2015, 15, 649–655.
- 16
- 16aN. Ramakrishnan, P. B. Sunil Kumar, J. H. Ipsen, Biophys. J. 2013, 104, 1018–1028;
- 16bM. Simunovic, C. Mim, T. C. Marlovits, G. Resch, V. M. Unger, G. A. Voth, Biophys. J. 2013, 105, 711–719.
- 17D. J. Kauert, T. Kurth, T. Liedl, R. Seidel, Nano Lett. 2011, 11, 5558–5563.
- 18
- 18aA. Kaufmann, V. Beier, H. G. Franquelim, T. Wollert, Cell 2014, 156, 469–481;
- 18bJ. B. Manneville, J. F. Casella, E. Ambroggio, P. Gounon, J. Bertherat, P. Bassereau, J. Cartaud, B. Antonny, B. Goud, Proc. Natl. Acad. Sci. USA 2008, 105, 16946–16951.
- 19H. Zhao, A. Michelot, E. V. Koskela, V. Tkach, D. Stamou, D. G. Drubin, P. Lappalainen, Cell Rep. 2013, 4, 1213–1223.
- 20J. C. Stachowiak, E. M. Schmid, C. J. Ryan, H. S. Ann, D. Y. Sasaki, M. B. Sherman, P. L. Geissler, D. A. Fletcher, C. C. Hayden, Nat. Cell Biol. 2012, 14, 944–949.
- 21A. N. Becalska, C. F. Kelley, C. Berciu, T. B. Stanishneva-Konovalova, X. Fu, S. Wang, O. S. Sokolova, D. Nicastro, A. A. Rodal, Mol. Biol. Cell 2013, 24, 2406–2418.
- 22M. R. Horton, S. Manley, S. R. Arevalo, A. E. Lobkovsky, A. P. Gast, J. Phys. Chem. B 2007, 111, 880–885.
- 23
- 23aB. Antonny, Curr. Opin. Cell Biol. 2006, 18, 386–394;
- 23bP. Sens, L. Johannes, P. Bassereau, Curr. Opin. Cell Biol. 2008, 20, 476–482.
- 24H. Yu, K. Schulten, PLoS Comput. Biol. 2013, 9, e 1002892.
- 25
- 25aH. Ewers, W. Romer, A. E. Smith, K. Bacia, S. Dmitrieff, W. Chai, R. Mancini, J. Kartenbeck, V. Chambon, L. Berland, A. Oppenheim, G. Schwarzmann, T. Feizi, P. Schwille, P. Sens, A. Helenius, L. Johannes, Nat. Cell Biol. 2010, 12, 11–18; sup pp. 11–12;
- 25bA. V. Shnyrova, J. Ayllon, I. I. Mikhalyov, E. Villar, J. Zimmerberg, V. A. Frolov, J. Cell Biol. 2007, 179, 627–633;
- 25cJ. Solon, O. Gareil, P. Bassereau, Y. Gaudin, J. Gen. Virol. 2005, 86, 3357–3363.