Quantitative Analysis of Location- and Sequence-Dependent Deamination by APOBEC3G Using Real-Time NMR Spectroscopy†
We thank the Ministry of Education, Science, Sports and Culture of Japan for Grants-in Aid for Scientific Research (24121714, 25115507, and 25291013 to M.K.; 23570146 and 24113710 to T.N.), Japan Science and Technology (CREST; M.K.), the Sumitomo–Denko and Iwatani Foundations (M.K.), and the Japan Society for the Promotion of Science (A.F.). For funding for open access charges, we are grateful to the Ministry of Education, Science, Sports and Culture of Japan (24121714).
Graphical Abstract
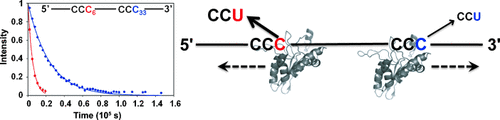
APOBEC3G (A3G) efficiently deaminates cytidines that are located close to the 5′ end of the single-stranded minus DNA of the HIV-1 genome. This process could be quantitatively analyzed using a newly developed real-time NMR spectroscopy method. As a result, the location-dependent deamination can be explained by two catalytic rate constants that depend on the direction of the approach to the target cytidine.
Abstract
The human antiretroviral factor APOBEC3G (A3G) deaminates the newly synthesized minus strand of the human immunodeficiency virus 1 (HIV-1), which results in the abolition of the infectivity of virus-infectivity-factor (Vif)-deficient HIV-1 strains.1–6 A unique property of A3G is that it deaminates a CCC hot spot that is located close to the 5′ end more effectively than one that is less close to the 5′ end. However, the mechanism of this process is elusive as it includes nonspecific binding of A3G to DNA and sliding of A3G along the DNA strand. Therefore, this process cannot be analyzed by existing methods using the Michaelis–Menten theory. A new real-time NMR method has been developed to examine the nonspecific binding and the sliding processes explicitly, and it was applied to the analysis of the deamination by A3G. As a result, the location-dependent deamination can be explained by a difference in the catalytic rates that depend on the direction of the approach of A3G to the target cytidine. Real-time NMR experiments also showed that A3G deaminates CCCC tandem hotspots with little redundancy, which suggests that A3G efficiently mutates many CCC hotspots that are scattered throughout the HIV-1 genome.