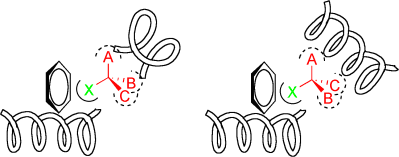Enantiocomplementary Enzymes: Classification, Molecular Basis for Their Enantiopreference, and Prospects for Mirror-Image Biotransformations
Paul F. Mugford Dr.
Department of Biochemistry, Molecular Biology & Biophysics and the Biotechnology Institute, University of Minnesota, 1479 Gortner Avenue, Saint Paul, MN 55108 (USA), Fax: (+1) 612-625-5780
Search for more papers by this authorUlrike G. Wagner Dr.
Department of Chemistry, University of Graz, Heinrichstrasse 28, 8010 Graz (Austria), Fax: (+43) 316-380-9840
Search for more papers by this authorYun Jiang
Department of Biochemistry, Molecular Biology & Biophysics and the Biotechnology Institute, University of Minnesota, 1479 Gortner Avenue, Saint Paul, MN 55108 (USA), Fax: (+1) 612-625-5780
Search for more papers by this authorKurt Faber Prof. Dr.
Department of Chemistry, University of Graz, Heinrichstrasse 28, 8010 Graz (Austria), Fax: (+43) 316-380-9840
Search for more papers by this authorRomas J. Kazlauskas Prof.
Department of Biochemistry, Molecular Biology & Biophysics and the Biotechnology Institute, University of Minnesota, 1479 Gortner Avenue, Saint Paul, MN 55108 (USA), Fax: (+1) 612-625-5780
Search for more papers by this authorPaul F. Mugford Dr.
Department of Biochemistry, Molecular Biology & Biophysics and the Biotechnology Institute, University of Minnesota, 1479 Gortner Avenue, Saint Paul, MN 55108 (USA), Fax: (+1) 612-625-5780
Search for more papers by this authorUlrike G. Wagner Dr.
Department of Chemistry, University of Graz, Heinrichstrasse 28, 8010 Graz (Austria), Fax: (+43) 316-380-9840
Search for more papers by this authorYun Jiang
Department of Biochemistry, Molecular Biology & Biophysics and the Biotechnology Institute, University of Minnesota, 1479 Gortner Avenue, Saint Paul, MN 55108 (USA), Fax: (+1) 612-625-5780
Search for more papers by this authorKurt Faber Prof. Dr.
Department of Chemistry, University of Graz, Heinrichstrasse 28, 8010 Graz (Austria), Fax: (+43) 316-380-9840
Search for more papers by this authorRomas J. Kazlauskas Prof.
Department of Biochemistry, Molecular Biology & Biophysics and the Biotechnology Institute, University of Minnesota, 1479 Gortner Avenue, Saint Paul, MN 55108 (USA), Fax: (+1) 612-625-5780
Search for more papers by this authorGraphical Abstract
Enzyme pairs that catalyze the same reaction but favor opposite enantiomers are known as enantiocomplementary enzymes (see scheme). To create mirror-image active sites, nature can switch the locations of binding sites and/or the locations of key catalytic groups. In this Minireview, X-ray crystal structures of enantiocomplementary enzymes are surveyed and classified into four groups.
Abstract
One often-cited weakness of biocatalysis is the lack of mirror-image enzymes for the formation of either enantiomer of a product in asymmetric synthesis. Enantiocomplementary enzymes exist as the solution to this problem in nature. These enzyme pairs, which catalyze the same reaction but favor opposite enantiomers, are not mirror-image molecules; however, they contain active sites that are functionally mirror images of one another. To create mirror-image active sites, nature can change the location of the binding site and/or the location of key catalytic groups. In this Minireview, X-ray crystal structures of enantiocomplementary enzymes are surveyed and classified into four groups according to how the mirror-image active sites are formed.
Supporting Information
Detailed facts of importance to specialist readers are published as ”Supporting Information”. Such documents are peer-reviewed, but not copy-edited or typeset. They are made available as submitted by the authors.
| Filename | Description |
|---|---|
| anie_200705159_sm_miscellaneous_information.pdf550.7 KB | miscellaneous information |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1Protein structures were compared by using the program DALI: L. Holm, J. Park, Bioinformatics 2000, 16, 566–567.
- 2R. C. L. Milton, S. C. F. Milton, S. B. H. Kent, Science 1992, 256, 1445–1448; commentary: G. Jung, Angew. Chem. 1992, 104, 1484–1486; Angew. Chem. Int. Ed. Engl. 1992, 31, 1457–1459.
- 3M. C. Fitzgerald, I. Chernushevich, K. G. Standing, S. B. H. Kent, C. P. Whitman, J. Am. Chem. Soc. 1995, 117, 11075–11080.
- 4B. Seelig, S. Keiper, F. Stuhlmann, A. Jäschke, Angew. Chem. 2000, 112, 4764–4768;
10.1002/1521-3757(20001215)112:24<4764::AID-ANGE4764>3.0.CO;2-V Google ScholarAngew. Chem. Int. Ed. 2000, 39, 4576–4579.10.1002/1521-3773(20001215)39:24<4576::AID-ANIE4576>3.0.CO;2-J CAS PubMed Web of Science® Google Scholar
- 5For a review, see: H. Griengl, A. Hickel, D. V. Johnson, M. Schmidt, C. Kratky, H. Schwab, Chem. Commun. 1997, 1933–1940; H. Griengl, H. Schwab, M. Fechter, Trends Biotechnol. 2000, 18, 252–256.
- 6K. Gruber, G. Gartler, B. Krammer, H. Schwab, C. Kratky, J. Biol. Chem. 2004, 279, 20501–20510.
- 7I. Dreveny, K. Gruber, A. Glieder, A. Thompson, C. Kratky, Structure 2001, 9, 803–815.
- 8I. Dreveny, C. Kratky, K. Gruber, Protein Sci. 2002, 11, 292–300.
- 9The methionine sulfoxides are epimers, not enantiomers, because they have the same configuration at Cα, but different configurations at the sulfoxide group. The Msrs show enantiocomplementary behavior with respect to reaction at the sulfoxide stereocenter. For a review, see: B. Kauffmann, A. Aubry, F. Favier, Biochim. Biophys. Acta Proteins Proteomics 2005, 1703, 249–260; for reports of enantioselectivity, see: V. S. Sharov, D. A. Ferrington, T. C. Squier, C. Schoneich, FEBS Lett. 1999, 455, 247–250; R. Grimaud, B. Ezraty, J. K. Mitchell, D. Lafitte, C. Briand, P. J. Derrick, F. Barras, J. Biol. Chem. 2001, 276, 48915–48920.
- 10W. T. Lowther, H. Weissbach, F. Etienne, N. Brot, B. W. Matthews, Nat. Struct. Biol. 2002, 9, 348–352.
- 11For reviews, see: R. J. Kazlauskas, A. N. E. Weissfloch, J. Mol. Catal. B 1997, 3, 65–72; T. Ema, K. Yamaguchi, Y. Wakasa, A. Yabe, R. Okada, M. Fukumoto, F. Yano, T. Korenaga, M. Utaka, T. Sakai, J. Mol. Catal. B 2003, 22, 181–192; for the example in Table 1, see: S. Vorlova, U. T. Bornscheuer, I. Gatfield, J.-M. Hilmer, H.-J. Bertram, R. D. Schmid, Adv. Synth. Catal. 2002, 344, 1152–1155; P. A. Fitzpatrick, A. M. Klibanov, J. Am. Chem. Soc. 1991, 113, 3166–3171; C. K. Savile, R. J. Kazlauskas, J. Am. Chem. Soc. 2005, 127, 12228–12229; see also Table S3 in the Supporting Information.
- 12M.-J. Kim, Y. I. Chung, Y. K. Choi, H. K. Lee, D. Kim, J. Park, J. Am. Chem. Soc. 2003, 125, 11494–11495.
- 13F. F. Huerta, A. B. E. Minidis, J.-E. Bäckvall, Chem. Soc. Rev. 2001, 30, 321–331.
- 14D. L. Ollis, E. Cheah, M. Cygler, B. Dijkstra, F. Frolow, S. M. Franken, M. Harel, S. J. Remington, I. Silman, J. Schrag, J. L. Sussman, K. H. G. Verschueren, A. Goldman, Protein Eng. 1992, 5, 197–211.
- 15P. F. Mugford, S. M. Lait, B. A. Keay, R. J. Kazlauskas, ChemBioChem 2004, 5, 980–987.
- 16M. Cygler, P. Grochulski, R. J. Kazlauskas, J. D. Schrag, F. Bouthillier, B. Rubin, A. N. Serreqi, A. K. Gupta, J. Am. Chem. Soc. 1994, 116, 3180–3186.
- 17S. Rhee, M. M. Silva, C. C. Hyde, P. H. Rogers, C. M. Metzler, D. E. Metzler, A. Arnone, J. Biol. Chem. 1997, 272, 17293–17302; R. A. John, Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1995, 1248, 81–96.
- 18D. Peisach, D. M. Chipman, P. W. Van Ophem, J. M. Manning, D. Ringe, Biochemistry 1998, 37, 4958–4967; S. Sugio, G. A. Petsko, J. M. Manning, K. Soda, D. Ringe, Biochemistry 1995, 34, 9661–9669; K. Yonaha, H. Misono, T. Yamamoto, K. Soda, J. Biol. Chem. 1975, 250, 6983–6989.
- 19S. Umhau, L. Pollegioni, G. Molla, K. Diederichs, W. Welte, M. S. Pilone, S. Ghisla, Proc. Natl. Acad. Sci. USA 2000, 97, 12463–12468; M. P. Simonetta, M. A. Vanoni, P. Casalin, Biochim. Biophys. Acta 1987, 914, 136–142.
- 20A. Mattevi, M. A. Vanoni, F. Todone, M. Rizzi, A. Teplyakov, A. Coda, M. Bolognesi, B. Curti, Proc. Natl. Acad. Sci. USA 1996, 93, 7496–7501; C. G. Mowat, A. Wehenkel, A. J. Green, M. D. Walkinshaw, G. A. Reid, S. K. Chapman, Biochemistry 2004, 43, 9519–9526.
- 21Z. X. Xia, F. S. Mathews, J. Mol. Biol. 1990, 212, 837–863.
- 22T. Fukui, N. Shiomi, Y. Doi, J. Bacteriol. 1998, 180, 667–673; Y.-M. Qin, M. H. Poutanen, H. M. Helander, A.-P. Kvist, K. M. Siivari, W. Schmitz, E. Conzelmann, U. Hellman, J. K. Hiltunen, Biochem. J. 1997, 321, 21–28.
- 23M. K. Koski, A. M. Haapalainen, J. K. Hiltunen, T. Glumoff, J. Biol. Chem. 2004, 279, 24666–24672.
- 24H. K. W. Kallwass, Enzyme Microb. Technol. 1992, 14, 28–35; W. Hummel, H. Schutte, M.-R. Kula, Appl. Microbiol. Biotechnol. 1985, 21, 7–15; H. Schutte, W. Hummel, M. R. Kula, Appl. Microbiol. Biotechnol. 1984, 19, 167–176.
- 25K. Niefind, H.-J. Hecht, D. Schomburg, J. Mol. Biol. 1995, 251, 256–281; U. Dengler, K. Niefind, M. Kiess, D. Schomburg, J. Mol. Biol. 1997, 267, 640–660.
- 26C. Vinals, E. Depiereux, E. Feytmans, Biochem. Biophys. Res. Commun. 1993, 192, 182–188.
- 27V. S. Stoll, M. S. Kimber, E. F. Pai, Structure 1996, 4, 437–447; K. Arai, T. Kamata, H. Uchikoba, S. Fushinobu, H. Matsuzawa, H. Taguchi, J. Bacteriol. 2001, 183, 397–400; T. Bhowmik, J. L. Steele, Appl. Microbiol. Biotechnol. 1994, 41, 432–439.
- 28P. D. Pawelek, J. Cheah, R. Coulombe, P. Macheroux, S. Ghisla, A. Vrielink, EMBO J. 2000, 19, 4204–4215; I. M. Moustafa, S. Foster, A. Y. Lyubimov, A. Vrielink, J. Mol. Biol. 2006, 364, 991–1002; see also: H. Zhang, M. Teng, L. Niu, Y. Wang, Y. Wang, Q. Liu, Q. Huang, Q. Hao, Y. Dong, P. Liu, Acta Crystallogr. Sect. D 2004, 60, 974–977; for biological properties, see: G. Ponnudurai, M. C. M. Chung, N.-H. Tan, Arch. Biochem. Biophys. 1994, 313, 272–378.
- 29O. Dym, E. A. Pratt, C. Ho, D. Eisenberg, Proc. Natl. Acad. Sci. USA 2000, 97, 9413–9418; M. Futai, Biochemistry 1973, 12, 2468–2474.
- 30D. S. Torok, S. M. Resnick, J. M. Brand, D. L. Cruden, D. T. Gibson, J. Bacteriol. 1995, 177, 5799–5805.
- 31D. R. Boyd, N. D. Sharma, S. A. Haughey, M. A. Kennedy, B. T. McMurray, G. N. Sheldrake, C. C. R. Allen, H. Dalton, K. Sproule, J. Chem. Soc. Perkin Trans. 1 1998, 1, 1929–1934.
- 32N. I. Bowers, D. R. Boyd, N. D. Sharma, P. A. Goodrich, M. R. Groocock, A. J. Blacker, P. Goode, H. Dalton, J. Chem. Soc. Perkin Trans. 1 1999, 1, 1453–1461.
- 33R. E. Parales, S. M. Resnick, C. Yu, D. R. Boyd, N. D. Sharma, D. T. Gibson, J. Bacteriol. 2000, 182, 5495–5504.
- 34A. Karlsson, J. V. Parales, R. E. Parales, D. T. Gibson, H. Eklund, S. Ramaswamy, Science 2003, 299, 1039–1042.
- 35N. H. Yennawar, M. E. Conway, H. P. Yennawar, G. K. Farber, S. M. Hutson, Biochemistry 2002, 41, 11592–11601; J. Davoodi, P. M. Drown, R. K. Bledsoe, R. Wallin, G. D. Reinhart, S. M. Hutson, J. Biol. Chem. 1998, 273, 4982–4989.
- 36J. Andexer, J. von Langermann, A. Mell, M. Bocola, U. Kragl, T. Eggert, M. Pohl, Angew. Chem. 2007, 119, 8833–8835; Angew. Chem. Int. Ed. 2007, 46, 8679–8681.
- 37C. Syldatk, O. May, J. Altenbuchner, R. Mattes, M. Siemann, Appl. Microbiol. Biotechnol. 1999, 51, 293–309; O. Keil, M. P. Schneider, J. P. Rasor, Tetrahedron: Asymmetry 1995, 6, 1257–1260.
- 38Y. H. Cheon, H. S. Kim, K. H. Han, J. Abendroth, K. Niefind, D. Schomburg, J. Wang, Y. Kim, Biochemistry 2002, 41, 9410–9417; J. Abendroth, K. Niefind, O. May, M. Siemann, C. Syldatk, D. Schomburg, Biochemistry 2002, 41, 8589–8597.
- 39F. P. Drijfhout, M. W. Fraaije, H. Jongejan, W. J. H. van Berkel, M. C. R. Franssen, Biotechnol. Bioeng. 1998, 59, 171–177.
10.1002/(SICI)1097-0290(19980720)59:2<171::AID-BIT5>3.0.CO;2-E CAS PubMed Web of Science® Google Scholar
- 40A. Mattevi, M. W. Fraaije, A. Mozzarelli, L. Olivi, A. Coda, W. J. van Berkel, Structure 1997, 5, 907–920.
- 41W. McIntire, D. J. Hopper, J. C. Craig, E. T. Everhart, R. V. Webster, M. J. Causer, T. P. Singer, Biochem. J. 1984, 224, 617–621.
- 42L. M. Cunane, Z. Chen, N. Shamala, F. S. Mathews, C. N. Cronin, W. S. McIntire, J. Mol. Biol. 2000, 295, 357–374.
- 43R. H. H. van den Heuvel, M. W. Fraaije, M. Ferrer, A. Mattevi, W. J. H. van Berkel, Proc. Natl. Acad. Sci. USA 2000, 97, 9455–9460.
- 44F. Oesch, Prog. Clin. Biol. Res. 1983, 135, 81–105.
- 45One exception is dl-2-haloacid dehalogenase, which displaces the halogen atom from both enantiomers with water at the same slow rate (<1 U mg−1): V. Nardi-Dei, T. Kurihara, C. Park, N. Esaki, K. Soda, J. Bacteriol. 1997, 179, 4232–4238. Racemases, single enzymes that convert both substrate enantiomers with approximately equal efficiency by using functionally symmetrical active sites, are another exception.
- 46S. A. Filppula, R. T. Sormunen, A. Hartig, W.-H. Kunau, J. K. Hiltunen, J. Biol. Chem. 1995, 270, 27453–27457.
- 47M. A. Phillips, M. R. Wildung, D. C. Williams, D. C. Hyatt, R. Croteau, Arch. Biochem. Biophys. 2003, 411, 267–276.
- 48R. Croteau, D. M. Satterwhite, D. E. Cane, C. C. Chang, J. Biol. Chem. 1986, 261, 13438–13445.
- 49I. A. Kaluzna, T. Matsuda, A. K. Sewell, J. D. Stewart, J. Am. Chem. Soc. 2004, 126, 12827–12832.
- 50D. Zhu, Y. Yang, L. Hua, J. Org. Chem. 2006, 71, 4202–4205; D. Zhu, C. Mukherjee, J. D. Rozzell, S. Kambourakis, L. Hua, Tetrahedron 2006, 62, 901–905.
- 51P. Cernuchova, M. D. Mihovilovic, Org. Biomol. Chem. 2007, 5, 1715–1719.
- 52D. E. Robertson, J. A. Chaplin, G. DeSantis, M. Podar, M. Madden, E. Chi, T. Richardson, A. Milan, M. Miller, D. P. Weiner, K. Wong, J. McQuaid, B. Farwell, L. A. Preston, X. Tan, M. A. Snead, M. Keller, E. Mathur, P. L. Kretz, M. J. Burk, J. M. Short, Appl. Environ. Microbiol. 2004, 70, 2429–2436.
- 53M. Chen-Goodspeed, M. A. Sogorb, F. Wu, F. M. Raushel, Biochemistry 2001, 40, 1325–1331; M. Chen-Goodspeed, M. A. Sogorb, F. Wu, F. M. Raushel, Biochemistry 2001, 40, 1332–1339.
- 54J. L. Vanhooke, M. M. Benning, F. M. Raushel, H. M. Holden, Biochemistry 1996, 35, 6020–6025.
- 55Y. Li, S. D. Aubert, E. G. Maes, F. M. Raushel, J. Am. Chem. Soc. 2004, 126, 8888–8889.
- 56S.-I. Ozaki, P. R. Ortiz de Montellano, J. Am. Chem. Soc. 1994, 116, 4487–4488; M. I. Savenkova, S. L. Newmyer, P. R. Ortiz de Montellano, J. Biol. Chem. 1996, 271, 24598–24603.
- 57Y. Hirose, K. Kariya, Y. Nakanishi, Y. Kurono, K. Achiwa, Tetrahedron Lett. 1995, 36, 1063–1066.
- 58Y. Koga, K. Kato, H. Nakano, T. Yamane, J. Mol. Biol. 2003, 331, 585–592.
- 59M. Ivancic, G. Valinger, K. Gruber, H. Schwab, J. Biotechnol. 2007, 129, 109–122.
- 60S. Bartsch, R. Kourist, U. T. Bornscheuer, Angew. Chem. 2008, 120, 1531–1534;
10.1002/ange.200704606 Google ScholarAngew. Chem. Int. Ed. 2008, 47, 1508–1511.
- 61A. O. Magnusson, M. Takwa, A. Hamberg, K. Hult, Angew. Chem. 2005, 117, 4658–4661;
10.1002/ange.200500971 Google ScholarAngew. Chem. Int. Ed. 2005, 44, 4582–4585.
- 62R. Sakowicz, M. Gold, J. B. Jones, J. Am. Chem. Soc. 1995, 117, 2387–2394.
- 63O. May, P. T. Nguyen, F. H. Arnold, Nat. Biotechnol. 2000, 18, 317–320.
- 64M. T. Reetz, B. Brunner, T. Schneider, F. Schulz, C. M. Clouthier, M. M. Kayser, Angew. Chem. 2004, 116, 4167–4170;
10.1002/ange.200460272 Google ScholarAngew. Chem. Int. Ed. 2004, 43, 4075–4078; M. D. Mihovilovic, F. Rudroff, A. Winninger, T. Schneider, F. Schulz, M. T. Reetz, Org. Lett. 2006, 8, 1221–1224.
- 65S. A. Funke, A. Eipper, M. T. Reetz, N. Otte, W. Thiel, G. Van Pouderoyen, B. W. Dijkstra, K.-E. Jaeger, T. Eggert, Biocatal. Biotransform. 2003, 21, 67–73.
- 66C. K. Savile, R. J. Kazlauskas, J. Am. Chem. Soc. 2005, 127, 12228–12229.
- 67D. Zha, S. Wilensek, M. Hermes, K. E. Jaeger, M. T. Reetz, Chem. Commun. 2001, 2664–2665.
- 68Y. Terao, Y. Ijima, K. Miyamoto, H. Ohta, J. Mol. Catal. B 2007, 45, 15–20.