Point-of-Care Testing for the Detection of MicroRNAs: Towards Liquid Biopsy on a Chip
Abstract
Point-of-care (PoC) testing is revolutionizing the healthcare sector improving patient care in daily hospital practice and allowing reaching even remote geographical areas. In the frame of cancer management, the design and validation of PoC enabling the non-invasive, rapid detection of cancer markers is urgently required to implement liquid biopsy in clinical practice. Therefore, focusing on stable blood-based markers with high-specificity, such as microRNAs, is of crucial importance. In this work, we highlight the potential impact of circulating microRNAs detection on cancer management and the crucial role of PoC testing devices, especially for low-income countries. A detailed discussion about the challenges that should be faced to promote the technological transfer and clinical use of these tools has been added, to provide the readers with a complete overview of potentialities and current limitations.
1 Introduction
In the era of precision medicine, liquid biopsy (LB) is reshaping the diagnostic and prognostic toolbox of clinical oncology providing multimodal methods for the non-invasive detection of cancer biomarkers in biological fluids, especially blood and urine.1, 2 By tracing the presence of circulating cells, extracellular vesicles, cell-free DNA or RNA (cf-DNA, cf-RNA), microRNA (miRNA), messenger RNA (mRNA) and proteins, LB is rewriting the standards of cancer management with a tangible economic impact in both low- and middle-income countries (LMICs) and high-income countries (HICs)3-5 contributing to improve the life quality of cancer patients.6 The impact of LB appears more relevant when numbers of cancer diagnosis and deaths are considered: in 2020, 19.3 million new cases and 10 million cancer-related deaths were recorded worldwide,7 while in 2023 ca. 2 million new cancer cases and approximately 600 k cancer deaths are estimated to occur only in the United States.6 More than 28 million new cancer cases are projected to occur in 2040, which corresponds to +47 % with respect to cases registered in 2020. As reported in Figure 1, this increase is expected to double the cases in low human development index (HDI) countries, from 0.67 to 1.25 million (+95 %), while in terms of the absolute burden, the high HDI countries are characterized by the maximum increase of cases, from ca. 7.4 to 11. 5 million new cases (+56 %).
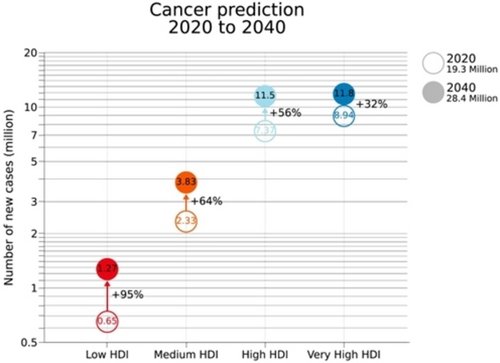
Projected number of new cases for all cancers combined (male and female have been considered) in 2040 according to the HDI (Copyright permission n. 5594710400458).7
Current projections assume a constant cancer incidence and mortality rate as of 2020 and are purely based on the expected growth and aging of the global population. The latter one is expected to reach 9.2 billion with the over 65 cohort likely to approach 25 percent of the total population by 2040. Therefore, it must be considered that these numbers might further increase due to occurring exposure to risk factors (e.g., smoking, western diet, high body weight).7 In this regard, the barriers towards high-quality cancer prevention and early detection might be lowered with the introduction of novel tumor specific biomarkers.
Among all biomarkers, miRNAs have attracted an increasing attention in the last decade. Since their discovery in the early 90s, the key role of miRNAs in the regulation of several processes from immune system stimulation to fat metabolism, cell proliferation and apoptosis, was described.8-10 The miRNAs are single-stranded noncoding RNAs of ca. 20 nucleotides that are pivotal regulators of gene expression.11 The main mechanism of action of miRNA relies on their binding to specific miRNA-binding sites on the targeted mRNA, mostly represented by the 3′untranslated region (3′UTR) of the transcript. In most cases, the miRNA-mRNA interaction results in the repression of the mRNA translation and its degradation, thus leading to the post-transcriptional silencing of targeted mRNA.12 Nevertheless, some miRNA-mRNA interactions enhance the stability of the mRNA promoting the translation of the transcript.13 The miRNA can also directly bind to DNA and interfere with the transcription of targeted genes.14 Aberrant levels of specific miRNAs were found to be associated with the incidence of numerous diseases from autoimmune syndromes to diabetes and cancer suggesting the potential applicability of miRNAs as therapeutics and biomarkers.15, 16 An overview of the main miRNAs action paths, their mode of secretion in the body and detection approaches is presented in Figure 2.
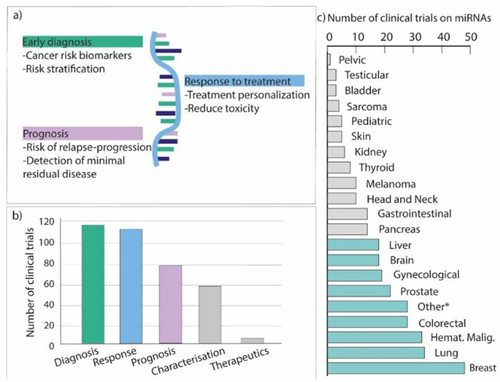
(a) Visual summary of the main clinical applications of miRNAs in relation to cancer growth, therapies, and diagnosis/prognosis. (b) Number of registered clinical trials in all Countries by ClinicalTrails.gov entailing the analysis of miRNA for the different clinical applications (i.e., diagnosis, response, prognosis, miRNA characterization and miRNA as a therapeutic tool). (c) Number of clinical trials investigating miRNA classified according to the tumor type. (Other *: pan-cancer studies (n=18), adrenocortical tumors (n=2), germ-cell tumors (n=1), endothelium tumors (n=1), midgut neuroendocrine tumors (n=1), urothelial carcinoma (n=1), seminoma (n=1), thymic carcinoma (n=1)); in light blue the cancer types with more than 20 clinical trials.
Compared to other circulating nucleic acid, miRNAs were found to be resistant to RNase activity. In fact, they show a remarkable stability in biological fluids even under harsh pH and after multiple freeze-thaw cycles.17, 18 miRNAs are thermally preserved via complexation with RNA-binding proteins or inclusion into extracellular vesicles like exosomes, but even free, circulating miRNAs show a great stability at room temperature for up to 24 h.19 These features make miRNAs suitable species for the development of detection tools. Clinically relevant information can be achieved by monitoring the variations in the expression levels of miRNAs in relation to the evolution of the disease and other aspects. The technological advances in the extraction and detection strategies for miRNAs as well as the possibility to screen them via PoC tests is contributing to increase their impact on the health sector.20, 21 In cancer management, the tissue-specific nature of miRNAs is allowing for the accurate identification of cancer types, sub-types, and primary origin of metastatic cancers.22 The impact of miRNAs-based LBs in cancer diagnosis and prognosis can be further enhanced by multiplex screening of a panel of several miRNAs or multiple circulating biomarkers (mRNA, miRNA, and long noncoding RNA (lncRNA)). In this work, benefiting of the recent advances in clinical trials for miRNAs identification, we discuss the potential impact and crucial role of PoC testing devices for miRNA-based LBs in relation to cancer detection and management. Moreover, we focused on the challenges that should be faced to promote the technological transfer and clinical use of these tools, with the aim of providing the readers a complete overview of potentialities and current limitations. Particular emphasis has been devoted to the electrochemical and colorimetric PoC testing devices, as most of the analytical approaches developed for miRNAs′ detection rely on these technologies.
2 miRNAs in Clinical Trials
Due to their complementary role in clinical practice to implement early cancer detection, to predict response to treatment and to refine the currently available prognostic algorithms, the interest around miRNAs in clinical oncology has been witnessed by the number of clinical trials that are or have been performed.23 Since 2001, 322 clinical trials involving miRNA analysis related to several human malignancies have been registered worldwide in ClinicalTrial.gov. Most of the registered clinical trials have been focused on, or have included miRNA characterization in the context of breast cancer, lung cancer, colorectal cancer, and hematological malignancies (Figure 2b–c). In this section, the most relevant clinical trials focused on the role of miRNAs in diagnosis, prediction and prognosis for clinical oncology practice are highlighted, both considering the specific tumor, the number of cohorts involved in the trial and their potential impact. 113 out of 322 (35 %) registered clinical trials were focused on the application of miRNA as a diagnostic supportive tool in clinical practice.
Lung cancer (LC) is the most frequently diagnosed type of cancer and represents the first leading cause of cancer-related death worldwide.7 Nowadays, computed tomography (CT) is the standard approach for LC screening in high-risk population (i.e., heavy smokers), but display a false positive rate that could lead to over-diagnosis.24 miRNA expression profiles in tumors tissue are indicative of aggressive LC development and specific miRNA signatures can be identified in plasma samples of patients up to two years before the clinical diagnosis of the tumor. BIOMILD is a prospective clinical trial involving 4,000 smoking volunteers that aims primarily at assessing the efficacy of plasma miRNA profiling as a first line screening test for LC detection in a high-risk population. Notably, an innovative microfluidic system coupled with a custom bioinformatic approach is exploited to interrogate the expression of a panel of 24 miRNAs of diagnostic relevance in LC (NCT02247453). The system was based on a custom-made Taq Assay RT-qPCR microfluidic card, which allowed the simultaneous quantification of 24 different miRNAs in plasma. The differential expression of miRNA was compared to the historic baseline miRNA quantification from the same patients and cut-off expression levels were computed to provide an accurate stratification of cancer risk (high, intermediate, and low).25
With regards to breast cancer (BC), the most common tumor in female, the ANDROMEDA study was performed as a prospective cohort-study, which involved 21,000 women attending BC screening, aimed at identifying women with high-risk to develop BC according to specific risk factors, including miRNA screening. To this end, the diagnostic accuracy of a panel of selected miRNAs associated to BC risk has been assessed by means of RT-qPCR analysis in LB samples to implement the optimal selection of high-risk patients (NCT02618538).26 In addition, to enhance the efficacy of early diagnosis towards BC, a multi-biomarker tool combining miRNA and ctDNA detection, namely Onco-liq, has been characterized, and it is currently under evaluation (estimated end: August 2023) on a cohort of 500 female patients in the 50–70 y/o range. Patients involved in the trials have not linked to oncological pathology, and the pilot is aimed to give the accuracy for BC early diagnosis and the management of positive patients. (NCT04906330).
In addition to early diagnosis, the early identification of predictors to follow treatment response are crucial to personalize therapeutic interventions and to understand the mechanisms leading to disease progression and therapeutic efficacy. The identification of predictive biomarkers of response and/or toxicity is particularly insidious, as it entails the thorough understanding of the pharmacodynamics and the pharmacokinetics of the administered drugs.27 109 out of 322 (34 %) clinical trials have investigated the potential predictive role of miRNA in term of treatments efficacy and safety. In locally advanced BC, the standard care relies on the neoadjuvant treatment followed by surgery. The early identification of patients, who can benefit from neoadjuvant chemotherapy, is crucial to select candidates for conservative surgical approaches. The predictive role of a 5-miRNA panel was recently evaluated in an initial pilot cohort of 120 BC patients receiving neoadjuvant treatment in the framework of an observational clinical trial (NCT01722851). Circulating miRNAs were shown to correlate with the likelihood of achieving a pathological complete response for BC therapies.28
Another example is represented by LIBIL, a prospective observational clinical trial that involves 900 patients affected by advanced non-small cell lung cancer (NSCLC), and it is aimed at evaluating the predictive role of miRNAs expression in plasma as an epigenetic factor associated to treatments response. The study is designed to involve patients without druggable mutations who are scheduled for chemotherapy (cohort 1), patients with oncogene-addicted disease treated with targeted therapy (cohort 2), and patients receiving immunotherapy (cohort 3) (NCT02511288). Most NSCLC patients are diagnosed at a late stage, when curative strategies are no longer possible. Therefore, choosing the optimal therapeutic strategies and identifying the leading mechanisms of drug resistance is crucial. Moreover, the possibility of interrogating miRNAs in a low-invasive manner would implement the tumor genome characterization when tissue biopsy is not amenable.
Depending on the role that miRNAs play in modulating the epithelial to mesenchymal transition, and tumor microenvironment, and the substantial role in tumor spread and metastatization, the prognostic implication of miRNAs analysis is currently under investigation in 74 out of 322 (23 %) clinical trials.29 For instance, IMMUNOREACT1 involves 466 participants affected by locally advanced rectal cancer (LARC) and it is aimed to characterize the tumor microenvironment, with the final goal of developing a prognostic test to predict nodal metastasis (NCT04915326). The latter will comprise the miRNA expression in the normal and health mucosa. The development of an immunological score specific for rectal cancer patients could refine the currently available risk stratification algorithms and help identifying patients who could benefit from organ sparing strategies or from more intensified chemoradiation treatments.30 The emerging potential role of miRNAs for diagnosis, prediction and prognosis is truly exciting, and the amount of ongoing clinical trials represents a clear signal that the near future will be, with good accuracy, characterized by novel established molecular benchmarks in the management of cancer. However, there are still several challenges to face, which concern bringing miRNA biomarkers in routinary LBs practice, such as miRNA standardization, the correction of technical and biological factors with respect to a reference miRNA sequence or another compound (e.g., creatinine in urine) as well as the unambiguous interpretation of the biological meaning of the alternation of miRNA levels. Among biological fluids, urine, saliva, and sweat should be favored in PoC testing design as they allow a non-invasive sampling, although miRNAs levels are often lower compared to blood samples. Therefore, the latter ones are the samples of election for LBs so far.31
3 PoC testing for miRNAs detection
Since pregnancy tests, blood glucose analyzers, and haemoglobin tests, as well as SARS-CoV-2 test kits, entered the market, the benefits and economic impact of PoC testing, also known as “near-patient testing”, became evident. In 2022, the global market for PoC diagnostics was valuated around $ 45 billion and it is expected to keep growing and reach $ 75 billion within the next five years.32 The investments in PoC aim at improving home-testing, with a focus on infections and autoimmune diseases. Based on the current existing PoC testing devices, and on the latest research within the field of liquid biopsy on chip, most of the approaches developed for miRNA detection relies on electrochemical and colorimetric tests (Figure 3).

(a) Electrochemical biosensor for miRNA-21 based on an amperometric readout adapted from Ref. [33]. (b) Example of commercial PGM. (c) PoC biosensor for the detection of miRNA-21 in urine obtained combining functionalized magnetic beads and PGM adapted from Ref. [34]. (Copyright permission n. 55946915111809). (d) Example of LFA for COVID-19 screening. (e) LFST with a dual readout including near-infrared-powered nanomotors for the detection of miRNA-21, adapted with permission from Ref. [35]. Copyright 2023 American Chemical Society. (f) Comparison of the discussed PoC for miRNAs in terms of costs (symbol: coins), sample values (amount of sample required; symbol: drop), user-friendliness (symbol: thumb up) and portability in a scale from 1 to 5 (symbol: hand bag). For portability and user-friendliness, 1 is poor and 5 optimal. For costs and sample amount, 1 correspond to a low value and 5 to a high one.
The main advantage related to these two signal transductions are: the absence of interferences from colored/turbid solution for electrochemical ones and the potential naked-eye visualization for colorimetric ones.36, 37 With respect to the electrochemical devices, the most utilized architecture for miRNA sensing is represented by complementary DNA, peptide nucleic acid (PNA) or RNA single strands that are chemically bound to a portable strip.38, 39 As described by the pioneering protocol of Plaxco et al., the use of recognition oligonucleotides modified with methylene blue (MB), a redox mediator, has been applied towards the detection of multiple miRNAs in biological fluids.40 A recent example has been reported concerning the prostate cancer, which describes the electrochemical detection of miRNA-21 in urine samples using a gold screen-printed electrode with limit of detection in the low nanomolar range, demonstrating a promising tool for cancer management.33 The strength of this architectures relies on the possibility to have all the system, both the recognizing oligonucleotides and the redox mediator, i.e. MB, immobilized on the portable device. Instead of MB, the recognition oligonucleotides have been also labeled with an enzyme, and this strategy has allowed to amperometrically detect miRNA-21 down to ca. 3 pM (Figure 3a).41 However, enzyme labelling might limit the platform sensitivity due to the steric hindrance, whereas the enzymatic activity could be affected by the presence of interfering compounds (e.g., inhibitors, competitive substrates, etc.) in the real matrices, leading to false negative/positive results. Another effective procedure for nucleic acid sensing was developed by the group of Shana Kelley: the oligonucleotide probes were not marked with MB, thus reducing the cost of probe production. Instead, an external redox mediator (ruthenium-based) was added for signal transduction.42 The assay exhibited excellent sensitivity and specificity in the detection of mutated ctDNA, down to 1 fg/μL of a target mutation in LC and melanoma patients, corresponding to detecting mutations at a level of 0.01 % relative to wild type. The same principle has been applied towards the detection of miRNA-492 in serum samples as a biomarker for pancreatic ductal adenocarcinoma diagnosis, using a PNA sequences as recognition probes.43 Although the use of an external redox mediator for transduction has the advantage of reducing the cost of the recognition element, the application of PoC is more complex, as it involves additional steps (multi-step sample pretreatment, etc.).
In addition to these examples, other innovative applications of electrochemical PoC towards miRNA detection have been described, which take benefits of the benchmark of the PoC, the personal glucose meters (PGMs), as showed in Figure 3b. In fact, the PGM electrochemical readout44-47 is living a second life as it enables the detection of non-glucose targets ranging from metal ion and small molecules to pathogens, proteins, and miRNAs.48, 49 PGMs have been coupled with biosensors for miRNAs by means of different recognition elements (e.g., invertase and amylase) and signal amplification methods (e.g., nanomaterials, hydrogels, DNAzyme) while employing glucose as signal tracer:50-52 the combination of DNAzyme and invertase has allowed the miRNA-21 detection down to 68 fM in urine samples (Figure 3c).34 In this case, the recognition oligonucleotides were immobilized on magnetic beads allowing to preconcentrate the target, to remove possible interferences and to prevent nonspecific adsorption by simply performing adequate washing steps of the beads (after incubation with the sample and prior to run the measurements). Therefore, compared to other enzymatic platforms, the ones relying on functionalized beads show an improved reliability, stability, and reproducibility, although they are characterized by multiple steps that might result in less user-friendly devices. An additional example demonstrating the efficacy of commercial PGM has been applied towards the detection of circulating miRNA-155: this target was quantified down to sub fM levels performing a dual-cascaded isothermal amplification reaction at 96-well plates, resulting in a surface-confined enrichment of invertase enzymatic probe catalyzing glucose production.53 Despite the limited instrumentation required by this strategy, the whole architecture is rather complex, resulting in analytical protocols inaccessible to non-expert. Similar experimental limitations characterize another strategy that has being highlighted in literature for miRNA determination, namely CRISPR/Cas.54 A duplex-specific nuclease-assisted CRISPR/Cas12a strategy has been reported for the simultaneous electrochemical determination of two sequences, miRNA-21 and miRNA-205.55 Here, the promising technology of CRISPR/Cas system has been combined with a magnetic bead-based electrochemical assay allowing to quantify the target sequences in the pM range with single-based mismatch selectivity. The detection miRNA-21 down to the low pM was also achieved using a rapid (<2 h) DNA-based amplification system relying on a toehold-mediated strand displacement reaction module (TDR) nanomachine coupled with a PGM.56 However, even if the adoption of commercial PGMs is more promising, if compared in terms of costs, portability, sample values and user-friendliness, than gold standard RT-qPCR (Figure 3f), the architectures are quite complex for non-specialized users. An analogous scenario can be observed when considering lateral flow assays (LFAs), which are widely used for pregnancy and COVID testing, assessment of food safety and detection of oncological and disease biomarkers (Figure 3d).57, 58 For the detection of miRNAs in serum, several lateral flow nucleic acid biosensors (LFNABs) have been reported.58, 59 Here, the capture of the miRNAs occurs mainly via sandwich-type nucleic acid hybridization reactions. The generated probe-target complex is conjugated with metallic nanoparticles and the visualization is usually carried out through different strategies, including colorimetry and photoluminescence.60 The clinical use of such LFNABs was first limited by time-consuming and expensive sample pre-treatment requiring miRNAs extraction and sequence amplification steps. The researchers cope with these limits by including signal amplification steps (i.e., catalytical hairpin assembly,61 oligonucleotide-templated reaction,62 rolling circle amplification,63 a wide variety of hybrid nanomaterials64-67 and even thermal treatments68). A LFNAB combined with a DNA barcode detection was recently applied in spiked human serum for the highly sensitive and selective multiplexing of exosomal miRNAs associated with colorectal cancer.69 As reported in the literature, although the RE-ASSURED criteria are often considered, the sensing performance of LFA-based devices were followed by a progressive complexification of the analytical architectures, resulting in a slow technological transfer and clinical applicability.70 To address the “complexification” issue, Chen et al. recently contributed to invert this trend by reporting an amplification-free LFNAB allowing for miR-21 detection in cell medium and spiked serum (Figure 3e).35 By introducing nanomotors probes, particles capable of self-motion upon NIR irradiation, the authors increase the capture efficiency of the target sequence of about 30 % within a simple, dual-mode LFA. It should be noted that, even if the literature is characterized by plenty of sensing architectures to be applied for miRNA sensing within the cancer field,71-74 the definition of PoC is strictly related to its efficacy close to/or near the patient.
Today, both electrochemical and LFA-based tests represent the main strategies to meet the need for user-friendly, cost-affordable, portable, and miniaturized devices, especially when compared with RT-qPCR (Figure 3f).20 In addition to these features, the possibility to perform multiplexing analysis can be achieved by designing different probes (to detect multiple miRNAs at the same platform) or by creating miniaturized arrays of sensors using integrated on-chip microfluidic systems (to detect panels of miRNAs in parallel channels). Apart for implementing the sensing devices, chemometric analysis and artificial intelligence-supported data elaboration should be introduced to verify the correlation between the different targets and acquire statistically relevant data (e.g., Ref. [75]).
In the following Table 1, the main features of the PoC testing devices described herein are summarized, which include target analyte, limit of detection (LOD), linear range, recovery %, samples nature, assay time. Also, with only one exception (see below), all the examples reported are proof-of-concept studies focusing on the development of analytical methods, which have not been validated on clinical samples yet. However, they have been tested in biological fluids spiked with known concentrations of miRNAs and further analyzed by means of RT-qPCR, as the gold standard methodology. Therefore, it can be expected that the next step would be the test with real samples followed by clinical deployment, if approved by the relevant Agencies (FDA, EMA).
Assay Type |
Target |
LOD |
Linear Range |
Recovery [%] |
Sample Matrix |
Average assay time |
Selective towards interfering DNA and RNA |
Ref. |
|---|---|---|---|---|---|---|---|---|
Voltammetric |
miRNA-21 |
2 nM |
50–340 nM |
90.0–105.2 |
Urine |
40 min |
Single-sequence miRNA |
[33] |
Amperometric based on PGM |
miRNA-21 |
68 fM |
100 fM–1 pM |
95.8–106.8 |
Urine of mice |
1 h 30 min |
n.a. |
[34] |
Optical and thermal LFTS |
miRNA-21 |
18 fM |
10 fM–1 pM |
n.a. |
Human serum and cell medium |
30 min |
Single-base mutated miRNA |
[35] |
Amperometric |
miRNA-21 |
29 fM |
0.096–25 pM |
n.a. |
Blood serum from BC patients; BC cells |
1 h |
Noncomplementary miRNA sequences |
[41] |
Voltammetric |
miRNA-492 |
6 nM |
50–100 nM |
97.6–102.2 |
Human serum |
50 min |
One-, two-base mismatches and scrambled sequences |
[43] |
Amperometric based on PGM |
miRNA-21, miRNA-335, miRNA-155, miRNA-122 |
0.325 fM (miR-21) |
0.5–250 fM (miR-21) |
n.a. |
Cellular extract |
40 min |
n.a. |
[50] |
Amperometric based on PGM |
miRNA-21 |
0.41 nM |
0.5–100 nM |
n.a. |
A549 cell lysate |
30 min |
miRNA-375, miRNA-141, miRNA-1290 |
[51] |
Amperometric based on PGM |
miRNA-21 |
19 pM |
50 pM–5.0 nM |
95–120 % |
Cell lysate |
1 h 20 min |
NoncomplementaryRNA sequences |
[52] |
Amperometric based on PGM |
miRNA155 |
0.36 fM |
1 fM–10 nM (dynamic range) |
97.7 % and 103.2 % |
Human serum
|
1 h 30 min |
miRNA21, miRNA141, miRNAlet-7a, miRNA155 |
[53] |
Amperometric based on PGM |
miRNA21, miRNA205 |
2.4 pM (miRNA-21), 1.1 pM (miRNA205) |
10 pM–100 nM |
98.5–102.5 % |
Fetal bovine serum and human serum |
1 h 45 min |
Noncomplementary RNA sequences |
[55] |
Amperometric based on PGM |
miRNA-21 |
200 pM |
n.a. |
n.a. |
Human serum |
2 h |
miRNAs and mutant DNA sequences |
[56] |
Visual LFNAB |
miRNA-21, miRNA-155, miRNA-210 |
0.007 nM (miRNA-155), 0.068 nM (miRNA-21), 0.017 nM (miRNA-210) |
0.01–5 nM (miRNA-155), 0.1–10 nM (miRNA-21), 0.05–10 nM (miRNA-210) |
n.a. |
Human serum |
10 min |
Single base-mismatched and two bases-mismatched DNAs |
[59] |
Visual LFA |
miRNA-21 |
0.89 pM |
5 pM–0.5 μM |
98.6–102.5 % |
A549, HeLa and HEK293T cells extracts and human serum |
55 min |
One-/three-bases mismatch RNA sequence |
[61] |
Fluorescence based LFA |
miRNA-150-5p |
9 nM |
10–200 nM |
n.a. |
Extracts from the plasma (n=18) |
15 min |
One-/two- base mismatch DNA sequences, miRNA-141 DNA |
[62] |
Colorimetric LFA |
miRNA 21, miRNA let-7a |
20 pM (miRNA let-7a), 40 pM (miRNA-21) |
20 pM–200 nM |
n.a. |
Human serum |
3 h |
Padlock probes (LP-21, LP-7a) and interference miRNA (miRNA 221) |
[63] |
Colorimetric LFA |
miRNA-21 |
0.3 pM |
1–50 pM |
n.a. |
Cell lysate |
15 min |
Random sequence, miRNA-155, miRNA-210, |
[64] |
Colorimetric LFA |
miRNA-21 |
1.0 pM |
1 pM–50 nM |
90.9–95.2 % |
Cancer cells extract and human serum |
20 min |
Non-complementary miRNA |
[65] |
Fluorescence based LFA |
miRNA-21 |
0.16 pM |
0.2–100 pM |
102–106 % |
Human serum |
2 h |
miRNAlet-7b and let-7d, miRNA-141, and miRNA-200a |
[67] |
Impedimetric |
miRNA-21, miRNA-155 |
From 1 and 200 ng/mL |
0.025–0.075 μg/mL |
n.a. |
Fetal bovine serum |
30 min |
Non-complementary and mismatch miRNA sequences |
[71] |
Voltammetric |
miRNA-21, miRNA-141 |
0.1 fM |
1 fM–1 nM |
97.3–108 % |
Human serum |
2 h |
Uric acid, miR-210 and dopamine |
[72] |
Voltammetric |
miRNA-21 |
5 pM |
5 pM–5 μM |
n.a. |
Human plasma |
10 min |
One-/two-bases mismatches RNA sequences |
[74] |
- PGM, personal glucose meter; LFTS, lateral flow test strip; LFNAB, lateral flow nucleic acid biosensors; LFA, lateral flow assay; n.a., not applicable.
4 Summary and Outlook
Early-stage cancer detection is of paramount importance for clinical application as it allows implementing effective and personalized strategies for therapeutic treatment of the patients. In this respect, LBs relying on miRNAs are particularly appealing as they can offer a minimally invasive method to assess the onset and progression of virtually any kind of cancer form. Although cutting-edge technologies are readily available in developed countries, the above-mentioned requirements might pose serious issues in developing countries, where even modestly expensive tests and basic facilities are often not available or missing. Furthermore, to translate LB applications into clinical routine, the standardization of protocols, along with the establishment of external quality control Schemes for each specific LB and the accreditation of laboratories that perform the assays is among the major challenges to be addressed.76 There is an urgent need to implement the PoC by developing new technologies, which are accurate, reliable, cost-effective, and portable.77 This implies that a novel multidisciplinary approach must be pursued, which must necessarily involve medical doctors, biologists, chemists, physicists, bioinformaticians and engineers (Figure 4a).
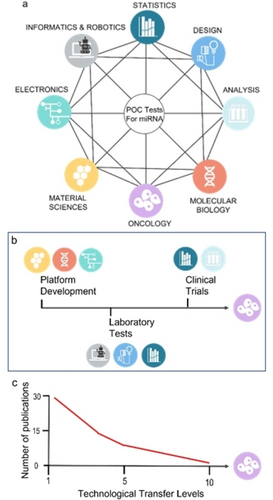
(a) A schematic representation of the network of disciplines required for the development and validation of PoC based on miRNAs showing the high degree of interconnections required. (b) Main steps of the Technological Transfer for PoC towards liquid biopsy application summarizing the disciplines involved in each step. (c) Number of publications (research articles, patents, scientific report) concerning PoC based on miRNAs in respect to their Technological Transfer Level (TTL).
Novel diagnostic strategies, like those exploited in the DNA chip array but specifically devised for miRNA, along with the advancement in bioengineering, material sciences, artificial intelligence/chemometrics and bioanalytical chemistry will allow the development of novel biosensing platforms fully integrated in miniaturized and portable devices.78, 79 Among others, those based on electrochemical and/or optical sensing platforms coupled with suitably designed microfluidics seem to be very promising to this end. Biosensing platforms relying on screen-printed electrodes, field effect transistor (FET),80, 81 surface plasmon resonance (SPR)75, 82, 83 or surface-enhanced Raman scattering (SERS)84-87 may offer the advantage of an unprecedented high sensitivity and fast detection times. Moreover, they can also be integrated with a second “dummy” sensing platform as the reference to provide a more accurate differential measurement. Label-free detection would be the preferred choice, as it avoids the use of too many reagents, thus decreasing the running costs. Whereas technologies based on electrochemical cells and FET would help lowering the production costs of the devices, as the optical components may be less prone to be miniaturized. Although the focus of the present work has been devoted to electrochemical and colorimetric PoC, we are aware of the potentialities and high analytical performances of the optical-based PoC (SERS and SPR). Therefore, we would like to suggest the readers some very interesting research work, which have been previously referenced.
On the other hand, with the aim of lowering the production costs, owing to highly sophisticated electronics and optical components, one should also think about employing a paper-based electrochemical platform.88 Paper offers several advantages: it is inexpensive and available everywhere; it can be easily patterned, employing wax or photoresistors, to provide microfluidic channels that help conveying the analyte in the measuring compartment; it naturally works as a filter, which is particularly interesting because it can easily filter out the corpuscles from complex matrixes such as blood; finally, conductive inks can be used to create electrode patterns of any dimension and fashion. To reach the clinic and bring LBs into cancer management routine, these technological advances should be followed by standardization processes establishing reliable control schemes for the use of PoC in miRNAs detection.76, 89 Therefore, the interdisciplinary network depicted in Figures 4b–c is essential not only for the technological transfer of PoC in LB, but also to make tangible advances in cancer management worldwide. The aim would be to provide economically affordable devices that are efficient, robust, accurate, sensitive, and user-friendly and that can be run at the point-of-care even by nonqualified personnel, as it already happens for the glucometer, and the pregnancy or the COVID-19 tests. A device that can also connect to a smart phone to provide in real time the results, which can be shared among medical doctors and analysts. This will surely lower the costs of the healthcare systems globally, thus allowing to invest resources for other activities for the common wealth.
Acknowledgments
The research leading to these results received funding from AIRC under MFAG 2022—ID. 27586 project—P. I. Stefano Cinti, and Ca’ Foscari University of Venice under Supporting Principal Investigator Grant 2021, TIDE project—P. I. Federico Polo. Giulia Moro acknowledges Fondazione Umberto Veronesi Postdoctoral Fellowship 2023.
Conflict of interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Biographical Information
Giulia Moro earned her joint Ph.D. from the University of Trieste (Italy) and the University of Antwerp (Belgium) under the supervision of Prof. K. De Wael and Prof. L. M. Moretto. Her Ph.D. was dedicated to the concept design of analytical methods for perfluoroalkyl substances monitoring. She subsequently received a postdoctoral fellow at the University of Venice. GM is currently a Fondazione Umberto Veronesi Fellow at the University of Naples, where she works on the design of biosensors for microRNA liquid biopsy.
Biographical Information
Chiara Dalle Fratte studied Pharmacy at the University of Padova (Italy) before moving to the National Cancer Center of Aviano for her Ph.D. in Biomedical Sciences under the supervision of Dr. Erika Cecchin. After a postdoctoral spell at the Venetian Institute of Oncology with Prof. Stefano Indraccolo, she moved to Germany where she currently is scientist at BioNTech in Mainz. She is also resident in Clinical Pharmacology at the University of Milan. Her scientific interests are liquid biopsy and personalized medicine.
Biographical Information
Nicola Normanno is the Director of the Translational Research Department of the IRCCS “Fondazione G. Pascale” of Naples. He is also President of the International Quality Network for Pathology (IQN Path) and President of the Italian Cancer Society (SIC). He has co-authored more than 300 publications in peer-reviewed journals, with an H-index of 80 and >30000 citations.
Biographical Information
Federico Polo earned a Ph.D. in Chemical Sciences in 2007 at the University of Padova. Afterwards, he spent several research stay abroad (USA and Germany) as postdoctoral fellow. In 2021 he was appointed associate professor at Ca’ Foscari University of Venice where he leads the Molecular Electrochemistry Laboratory. His research area focused on molecular electrochemistry, electron transfer mechanism and electrogenerated chemiluminescence in molecular model systems.
Biographical Information
Stefano Cinti is an Associate Professor at the Department of Pharmacy, University of Naples “Federico II”. He leads the Uninanobiosensors Lab, and his research includes the development of electrochemical sensors, portable diagnostics, paper-based devices, and nanomaterials. He has published more than 80 papers in peer-reviewed journals, with an H-index of 32 and >4000 citations. He has been awarded with a My First AIRC Grant to develop liquid biopsy on chip for Triple Negative Breast Cancer management. He loves communicating science to nonspecialized audiences through TV shows, radio, and books.









