In office sampling of eosinophil peroxidase to diagnose eosinophilic chronic rhinosinusitis
Abstract
Background
Practical biomarkers for endotypic characterization of chronic rhinosinusitis (CRS) remain elusive, hindering clinical utility. Eosinophil peroxidase (EPX) is an enzyme released by activated eosinophils. The objective of this study was to evaluate a clinic EPX assay as a marker of eosinophilic CRS.
Methods
Subjects with and without CRS presenting to a tertiary care rhinology clinic were prospectively enrolled, and nasal cytology brushings were collected from the middle meatus during in-clinic nasal endoscopy. ELISA assay was used to quantify EPX levels, and a customized multiplex immunoassay was used to quantify inflammatory cytokine mediators. Findings were correlated with clinical data.
Results
Forty-two subjects were enrolled, including 31 CRS subjects and 11 controls. Median EPX levels were 125.0 ng/mL (standard deviation [SD] 1745.8) and 6.5 ng/mL (SD 99.0) for CRS group and controls, respectively (p = 0.003). EPX levels were associated with history of asthma (p = 0.015), allergies (p = 0.028), polyps (p = 0.0006), smell loss (p = 0.006), and systemic eosinophilia or elevated immunoglobulin E (p ≤ 0.0001). Twenty-eight subjects from both the CRS and control groups had prior pathology for comparison, with histologic confirmation of local tissue eosinophilia (>10 eosinophils/hpf) in 11 subjects. This subgroup had a median EPX level of 967.5 ng/mL compared to 10.6 ng/mL in 17 subjects without local tissue eosinophilia (p = 0.0008). EPX levels were positively correlated to interleukin-5 levels (p = 0.0005).
Conclusion
EPX levels can be measured via well-tolerated in-clinic collection of nasal mucus. EPX levels are associated with clinical markers of type 2 inflammation and tissue eosinophilia and may provide a valuable diagnostic tool to delineate eosinophilic CRS.
1 INTRODUCTION
Endotypic characterization of chronic rhinosinusitis (CRS) is of great intellectual and investigative interest but is rarely employed in the clinic due to lack of diagnostic tools. The ability to discriminate inflammatory endotypes via in-clinic testing could have several benefits for patients and providers. Identifying patients with type 2 inflammation is particularly relevant given the recent availability of biologic therapies targeting type 2 inflammatory cascades.1 In-clinic testing could also allow for objective evaluation of disease progression and response to therapeutics. Widespread, real-time information on key inflammatory drivers of CRS-associated disease processes will encourage further development of targeted therapeutics.
Type 2 inflammation has been associated with eosinophilia and interleukin (IL) cytokine mediators, IL-4, IL-5, and IL-13, which contrasts non-type 2 inflammation. The latter categorization can be further delineated into types 1, 3, and unclassified endotypes, and have been associated with neutrophilia and cytokine mediators interferon-gamma (IFN-γ) and IL-17.2 Currently, the gold standard for diagnosis of type 2 inflammatory or eosinophilic CRS (eCRS) involves surgical tissue sampling with histologic enumeration of eosinophils per high powered field (hpf), with ≥10 eosinophils/hpf being a commonly used cutoff.3, 4 Because clinical diagnosis and treatment decisions must be made prior to reaching the point of surgery, our diagnostic acumen relies on clinical history and nasal endoscopic assessment to evaluate clinical phenotype, with polyps often used as a proxy for type 2 inflammation. This method is limited—a history of asthma and nasal polyps does not always correlate with type 2 inflammation, and patients without polyps on endoscopy have been found to have type 2 inflammation.5 This diagnostic challenge makes it difficult to individualize therapy, and likely contributes to the high rates of patients who do not respond to expensive type 2 targeted biologic therapies.6
Recent literature supports a potential role for eosinophil peroxidase (EPX), an enzyme localized in eosinophil granules, as a biomarker of type 2 inflammation in CRS. EPX a unique eosinophil granule protein in that it is one of only two eosinophil granule proteins exclusive to eosinophils, and it is the most abundant eosinophil granule protein within the eosinophil matrix.7 In the first study evaluating EPX in patients with CRS, EPX levels measured via tissue immunohistochemical staining were increased in patients with CRS compared to controls and correlated with markers of type 2 inflammation.8 Kobayashi et al. demonstrated lower protein phosphatase p = 0 (PP2A) activity in patients with CRS with nasal polyposis (CRSwNP), an effect that was intensified in the subgroup with severe asthma.9 Given a known pathway in which EPX phosphorylates and thus increases degradation of PP2A, this study indirectly suggested EPX levels may be elevated in this CRS endotype. Eosinophil cationic protein, a similar granule protein to EPX also released by activated eosinophils, has been shown to predict nasal polyposis recurrence post-operatively.10 In the last year, interest has grown for this exciting potential biomarker of eCRS. Smith et al. evaluated a novel EPX activity assay demonstrating increased EPX activity and ethmoid tissue EPX protein levels in patients with pathologically diagnosed eCRS.11 The authors also demonstrated an association between EPX activity and Lund‒Kennedy scores. Idler et al. demonstrated that higher EPX levels in ethmoid tissue corresponds to higher tissue eosinophilia in patients with CRS and was correlated with sinonasal outcomes test-22 (SNOT-22) and Lund‒Mackay scores.12 Taken together, these data support our hypothesis that sinonasal EPX can be utilized as a novel biomarker to identify eCRS, and it may be linked to increased disease severity in these patients. In this study, we aim to evaluate EPX in nasal mucus as a biomarker of type 2 inflammation by correlating protein levels to objective clinical markers of type 2 inflammation and to relative expression of inflammatory cytokines.
2 MATERIALS AND METHODS
2.1 Patient enrollment
This study was approved by the University of California San Francisco (UCSF) IRB (#11-07750).
Subjects were prospectively enrolled in this study from UCSF Sinus Center. All study group subjects had a clinical diagnosis of CRS based on 2015 Rhinosinusitis Task Force guidelines,13 including experiencing at least 2/4 of the following symptoms for >/ = 2 consecutive weeks (1) nasal congestion; (2) anterior or posterior mucopurulent drainage; (3) facial pain, pressure, or fullness; and (4) a decreased sense of smell as well as evidence of inflammation based on CT or endoscopic exam within 1 month of enrollment. All control group subjects were pre- or post-operative patients undergoing extended endoscopic approach to pituitary lesions, with no prior history of chronic sinonasal symptoms. These control subjects were a minimum of 6 weeks post-operative and had well-healed mucosa on endoscopic examination. Patients were excluded from participation if they were <18 years of age, had been taking oral prednisone within 2 weeks of clinic visit, had history of prior endoscopic sinus surgery within 1 year, were on biologic therapy, or had diagnosis of immunodeficiency, sinonasal malignancy, or autoimmune disease. Because our aim was to evaluate the diagnostic accuracy of EPX in delineating CRS due to type 2 inflammation, we also included patients unlikely to exhibit a type 2 inflammatory endotype, such as CRS due to cystic fibrosis or ciliary dysmotility syndromes.
2.2 Clinical data collection
All subjects underwent complete clinical evaluation, involving collection of medical history and sinonasal-specific history. This included demographic data, medical comorbidities, sinonasal surgical history, current sinonasal regimen, allergies, smoking status, and nonsteroidal anti-inflammatory drug sensitivity. Subjects completed the SNOT-22 as a part of standard clinical care.
2.3 Biospecimen collection, processing, and EPX protein level assay
Sinonasal specimens were obtained via nasal cytology brushings of the middle meatus under endoscopic guidance using ConMed bronchoscope sheathed 3.0 mm cytology brushes (CONMED Corporation). Each patient was swabbed twice within the same middle meatus. Each cytology brush head was placed in Dulbecco's phosphate-buffered saline (D-PBS). Specimen were immediately placed on ice and processed within 4 h of collection. Processing involved vortex and centrifugation to separate the specimen from the brush followed by removal of the D-PBS. The two specimen per patient were combined and weighed, and a 5× volume of D-PBS with 10% sputolysin was added. A series of 3 × 5-min warm water bath incubations at 37°C followed by mixing via pipette was performed to break down bonds of the eosinophilic mucin within which EPX resides. Cell counting was performed, and cell slides were made. The remaining specimen was centrifuged to separate supernatant and cell pellet, and these were stored at −80°C. When ready for assaying, the supernatant was thawed on ice and utilized to quantify EPX protein expression via EPX ELISA kit from Diagnostics Development.
2.4 Cytokine analysis
Using same supernatant collected for EPX protein quantification via ELISA, we used the Meso Scale Discovery Custom U-plex Assay Platform to quantify cytokine levels, including IFN-γ, IL-4, IL-5, IL-6, IL-9, IL-13, IL-17, IL-33, and thymic stromal lymphopoietin protein (TSLP). These cytokines were chosen given their roles in driving pathogenesis of CRS endotypes.2 Sixty-four percent of samples were run in duplicate, with the remaining samples run in a single well as limited by sample quantity. Per pilot testing performed to optimize detection of cytokines within the suggested fit curve range, samples were run in an undiluted fashion and incubated with the detection antibody solution overnight at 4°C.
2.5 Statistical analyses
Using a two-tailed alpha of 0.05 with our study powered to 80%, we were powered to detect a 50% higher EPX level in eosinophilic CRS patients as compared to non-eosinophilic counterparts with a total sample size of N = 32 samples (N = 16 eCRS patients and 16 non-eCRS patients). This was a conservative measure, as we hypothesized that even in the presence of some mixed inflammation, our non-eCRS group (reflecting <10 eosinophils/hpf) would have less than 50% of the EPX levels seen in the eCRS group. To further probe our diagnostic question, we added a “clean” control group, with the goal of recruiting 10 subjects without CRS, expected to demonstrate negligible levels of EPX. Comparisons between groups were performed with the assumption of non-parametric data (confirmed via Shapiro‒Wilks normality testing) using Wilcoxon rank sum testing, Spearman correlation, or Kruskal‒Wallis testing as appropriate based on variable type. Dunn's multiple comparisons test was used for inter-group comparisons if significance was identified on Kruskal‒Wallis testing. Linear regression analyses were performed to control for potential confounders. StataSE/version 17.0 (StataCorp.), Microsoft Excel version 16.79.1 (Microsoft Corporation, 2023), and GraphPad Prism version 10.0.0 (GraphPad Software) were used for statistical analyses and graphs.
3 RESULTS
3.1 Demographics
A total of 42 patients were enrolled in the study, 31 with a diagnosis of CRS and 11 non-CRS controls. Of the control cohort, seven (64%) had prior extended endoscopic approach for resection of a pituitary adenoma, two (18%) had prior endoscopic dacryocystorhinostomy, one (9%) had prior endoscopic medial orbital wall decompression, and one (9%) had prior endoscopic repair of an encephalocele. The mean age in years of all subjects was 56 (range of 20‒76). Thirty-eight percent of the cohort was female, and there was a higher proportion of females in the control group (p = 0.011, Table 1). Self-reported race and ethnicity also differed between groups with higher proportion of non-white patients (p = 0.018) and higher proportion of Hispanic or Latino patients (p = 0.0006) in the control group. Mean body mass index did not differ between groups (Table 1).
| Demographics | CRS (N = 31), N (%) | Non-CRS controls (N = 11), N (%) | p-value |
|---|---|---|---|
| Female sex | 8 (26) | 8 (73) | 0.011 |
| Mean age (range) | 56.7 (20‒76) | 53.6 (23‒68) | 0.439 |
| Self-reported race | 0.018 | ||
| White | 23 (74) | 5 (45) | |
| Asian | 6 (19) | 1 (9) | |
| Other | 1 (3) | 4 (36) | |
| Declined | 1 (3) | 1 (9) | |
| Self-reported ethnicity | 0.006 | ||
| Hispanic or Latino | 2 (6) | 5 (45) | |
| Not Hispanic or Latino | 28 (90) | 5 (45) | |
| Declined | 1 (3) | 1 (9) | |
| Mean BMI (SD) | 26.7 (4.6) | 28.9 (6.1) | 0.423 |
- Note: Bolded p-values indicate significant difference between groups (p > 0.05).
- Abbreviations: BMI, body mass index; SD, standard deviation.
3.2 Receiver operating characteristic curve for EPX level predicting eCRS
A subgroup of patients had pathology reports available from prior surgery, and these were utilized to evaluate whether EPX level could be utilized to predict eCRS. Patients with >10 eosinophils/hpf were considered to be eCRS, while the remaining patients with a diagnosis of CRS but <10 eosinophils/hpf were considered non-eCRS. Twenty-nine subjects had pathology available, including all 11 non-CRS controls and 18 CRS patients. Of the CRS patients, four had <10 eosinophils/hpf (non-eCRS) and 14 had pathology reports indicating >10 eosinophils/hpf (eCRS). Four of these patients had surgery performed shortly after in-clinic nasal mucus collection allowing for prospective evaluation of eosinophils/hpf in relation to EPX level. All four had elevated eosinophils/hpf (>/ = 60) on pathologic evaluation and a mean EPX of 2302 ng/mL (standard deviation [SD] 2471). The EPX levels were significantly higher in the eCRS group (mean 1570, SD 2364), compared to the non-eCRS (mean 19, SD 30) and the non-CRS control groups (mean 48, SD 99) (p = 0.001, Figure 1A).
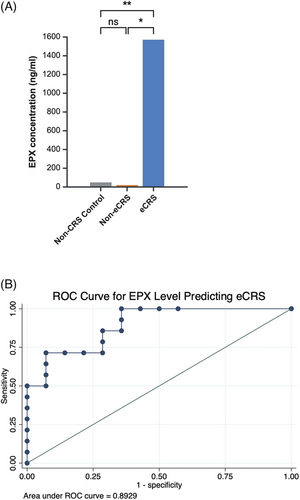
We performed receiver operating characteristic curve analysis to evaluate the ability to discriminate eCRS diagnosis based on EPX levels. Area under the curve (AUC) was 0.89, indicating a good level of discrimination between eCRS and non-eCRS cases (Figure 1B). The optimal cutoff point, determined using Youden's J statistic to identify a threshold at which sensitivity and specificity are balanced, is an EPX level of 118.35 ng/mL. At this threshold, the sensitivity is 71.4% and specificity is 92.9%.
3.3 High and low EPX groups
To evaluate associations between EPX and markers of type 2 inflammation, we grouped the CRS patients into “high” and “low” EPX groups based on the above cutoff of 118.35 ng/mL. In comparing the eCRS group to the high and low EPX groups, all 10 subjects with pathology data in the high EPX group were eCRS patients. There were also four subjects in the eCRS group who fell into the low EPX categorization. Notably, four patients had a diagnosis of either cystic fibrosis or ciliary dysmotility. The mean EPX level in this subgroup was 24 ng/mL (SD 40), and none of these subjects belonged to the high EPX group. Finally, we evaluated the cohorts as they relate to prior objective evidence of systemic type 2 inflammation. Twenty-four subjects were previously tested for serum immunoglobulin E (IgE) and/or had prior complete blood count with differential. Within the high EPX group, 10 of 11 (91%) subjects tested had a prior blood test showing elevated serum IgE or absolute eosinophil count (AEC), versus one of eight (13%) subjects in the low EPX group (p < 0.0001). Four patients in the non-CRS control group had prior testing, 0 of which showed elevated type 2 blood markers.
3.4 Cell slide data
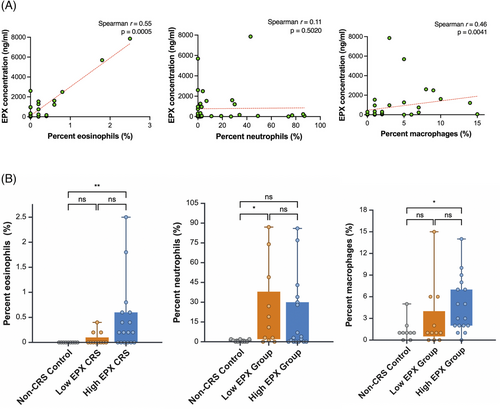
Cell counts for percent eosinophils, neutrophils, and macrophages were performed. EPX level in all subjects was positively correlated with percent eosinophils (Spearman r = 0.55, p = 0.0005) and percent macrophages (Spearman r = 0.46, p = 0.004, Figure 2A). Percent neutrophils were not correlated to EPX level (p = 0.502, Figure 2A). The subgroup with a diagnosis of CF or ciliary dysmotility had a mean percent neutrophil of 16.6% (SD 27.3), as compared to 29.3% in other non-eCRS subjects (SD 49.9, p = 1.0). Cell counts were also analyzed by EPX subgroups. The high EPX group demonstrated higher percent eosinophils than the non-CRS control group (means 0.46%, SD 0.69 and 0.0%, SD 0.0, respectively; p = 0.004). The high EPX group did not have a higher percent eosinophils than the low EPX group (mean 0.07%, SD 0.13, p = 0.08; Figure 2B). Percent neutrophils was higher in the low EPX group (mean 24.9%, SD 31) as compared to control (mean 0.8%, SD 1, p = 0.041). Percent macrophages was higher in the high EPX group (mean 4.8%, SD 3.9) compared to control (mean 1.3%, SD 1.5, p = 0.024). There was not a significant difference in percent macrophages in the high EPX compared to the low EPX group (mean 3.0%, SD 4.5, p = 0.12, Figure 2B).
3.5 Markers of type 2 inflammatory phenotype
Several clinical variables associated with type 2 inflammation were evaluated for associations with EPX levels in all subjects. EPX levels were correlated with history of asthma (p = 0.015) or allergies (p = 0.028), aspirin or NSAID sensitivity (p = 0.044), smell loss (p = 0.006), and history of elevated serum IgE or AEC (p < 0.0001). The median EPX level in subjects with nasal polyps on endoscopy was 847 ng/mL (IQR 58‒1535 ng/mL) versus 25 ng/mL (IQR 1‒79 ng/mL) in subjects without nasal polyps on endoscopy (p = 0.0006). Within the subjects with CRS only, higher EPX levels were also correlated with nasal polyps (p = 0.02). EPX level did not correlate with a history of atopic dermatitis or eczema (p = 0.953) or SNOT-22 score (p = 0.103). We also evaluated the EPX subgroups as they relate to these type 2-associated variables (Table 2). Purulence, a potential phenotypic marker for type 1 inflammation, was only found in the low EPX group (p = 0.010).
| Clinical history and presentation | High EPX CRS (N = 17), N (%) | Low EPX CRS (N = 14), N (%) | Non-CRS controls (N = 11), N (%) | p-value |
|---|---|---|---|---|
| History of asthma | 8 (47) | 5 (36) | 0 (0) | 0.017a |
| History of allergies | 11 (65) | 10 (71) | 3 (27) | 0.069 |
| History of atopic dermatitis | 1 (6) | 1 (7) | 0 (0) | 1.0 |
| Median SNOT-22 score(IQR) | 32 (12‒56) | 35.5 (15‒45) | 7.5 (0‒21) | 0.021a, b |
| Endoscopic exam findings | ||||
| Polyps | 13 (76) | 5 (36) | 0 (0) | <0.0001a |
| Purulence | 1 (6) | 6 (43) | 0 (0) | 0.006b, c |
| Allergic mucin | 4 (24) | 0 (0) | 0 (0) | 0.054 |
| Hyposmia or anosmia | 7 (41) | 1 (7) | 0 (0) | 0.010a |
| Aspirin or NSAID sensitivity | 2 (12) | 0 (0) | 0 (0) | 0.328 |
- Note: The high EPX CRS group represents CRS patients with EPX levels ≥118.35 ng/mL and the low EPX CRS group represents CRS patients with EPX levels <118.35 ng/mL. Bolded p-values indicate significant difference between groups (p > 0.05).
- Abbreviations: CRS, chronic rhinosinusitis; IQR, interquartile range; NSAID, non-steroidal anti-inflammatory; SNOT-22, sinonasal outcomes test-22.
- a Statistically significant difference (p < 0.05) on Dunn's multiple comparison test between the high EPX and control groups.
- b Statistically significant difference (p < 0.05) on Dunn's multiple comparison test between the low EPX and control groups.
- c Statistically significant difference (p < 0.05) on Dunn's multiple comparison test between the high EPX and low EPX groups.
Notably, a proportion of patients in all subgroups had a clinical history of allergies and, as noted above, higher EPX levels were associated with a history of allergies (p = 0.028). The median EPX level in subjects with allergies 105 ng/mL (IQR 37‒1088 ng/mL) versus 25 ng/mL (IQR 0‒161 mg/mL).
3.6 Inflammatory cytokines
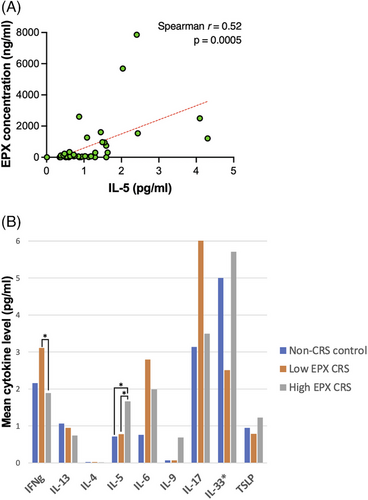
Cytokines levels (in pg/mL) were analyzed in comparison to EPX levels in all subjects. The only cytokine associated with EPX level was IL-5 (Spearman r = 0.52, p = 0.0005, Figure 3A). No significant relationship was identified between IFN-γ, IL-4, IL-6, IL-9, IL-13, IL-17, IL-33, or TSLP and EPX level (p > 0.05 for all cytokines). Cytokine levels within EPX subgroups were also analyzed (Figure 3B). IL-5 levels were higher in the high EPX group as compared to non-CRS control group (p = 0.01) and low EPX CRS group (p = 0.037). IFN-γ levels were higher in the low EPX CRS group as compared to the high EPX CRS group (p = 0.012). The remaining cytokine levels were not significantly different between EPX subgroups (p > 0.05 for all cytokines). Of note, despite optimization pilot studies and use of undiluted specimens, IL-4 was frequently below detectable limits of the test (16/42 subjects), and thus reported results for this cytokine are less reliable.
4 DISCUSSION
This study marks the first instance of implementing testing of EPX protein levels within nasal mucus collected in clinic, representing a novel proof-of-concept. Previously published studies have evaluated EPX levels in tissue homogenates from surgically collected tissue samples but have yet to validate EPX levels within nasal mucus collected via in-clinic nasal brushings.8, 11, 12 The EPX and cytokine levels in our samples represent those found in nasal mucus as opposed to within immune or epithelial cells. Although we were able to obtain cellular data from cytology brushings, our processing method separates cells from the secreted nasal mucus without disrupting the cell membranes. Our processing method may represent a more sensitive test of disease severity by specifically measuring the EPX released by degranulated eosinophils, as opposed to additionally measuring EPX stored within eosinophils. It is important to note that though our test is collected in clinic, which is an important step forward in the realm of endotyping, it does not yet represent a point-of-care (POC) test, as the collected specimen are then analyzed in the laboratory. It is feasible for this to be translated to standard patient care, similar to other tests collected in clinic and sent to a laboratory; however, a POC test could be of particular utility in the future and is another potential area for development.
Throughout the study, we categorized subjects with CRS into “high” and “low” EPX groups based on a predictive cutoff point determined using Youden's J statistic. When EPX level is above this defined threshold, the EPX test is considered positive, or representative of eCRS. While this statistical test allows us to determine the EPX level that represents the most balanced sensitivity/specificity for predicting eCRS status based on histopathology, it is limited by the data inputted. With higher numbers of subjects this would become increasingly accurate. Furthermore, some accuracy is lost given the tissue collected for pathology that this testing is based on was not performed simultaneously to the collection of nasal mucus. If EPX level is to be used as a diagnostic test in the future, the threshold at which its elevation truly represents eCRS must be more clearly established. It is likely that low EPX subjects with prior eosinophilic pathology had better controlled disease at the time of nasal brushing, resulting in less eosinophil degranulation and consequently lower EPX levels. We demonstrated that EPX level is also associated with a history of allergies, and this is perhaps why there are select individuals with non-zero EPX levels even within the non-CRS control group. This confounding factor will have to be controlled for when establishing an EPX threshold that represents true eCRS. This does however suggest potential utility of this diagnostic test to delineate allergic from non-allergic rhinitis.
An additional finding of interest was the correlation between EPX levels and macrophages on cell slide data. This is consistent with studies demonstrating increased level of macrophages within sinonasal tissue of polyp patients.14 Alternatively activated (M2) macrophages are implicated in type 2 inflammatory states, and have been positively correlated with IL-5 and eosinophil cationic protein in CRS patients.15
Contrary to our expectations, we did not find an association between SNOT-22 score and EPX levels, and this was true even within the high EPX group and the group with prior pathology confirming eCRS. We postulate that this is due to variance in baseline local eosinophilia; if patients have very high baseline and then improve, their EPX level may drop but still be quite high relative to other patients. Similarly, comparing a change in EPX level to change in SNOT-22 score may be more clinically meaningful. Given our current study only provides a single snapshot in time, we are not able to evaluate relative change in EPX within subjects, which may be a more valuable prognostic tool. Future studies quantifying changes in EPX levels over time and response to disease treatment will be critical to clarify the diagnostic and prognostic role of this protein.
We acknowledge that the demographic differences in sex, race, and ethnicity between our CRS and non-CRS control cohorts is a limitation of this study. As mentioned above, the lack of simultaneous tissue collection to confirm eosinophilic inflammation via eosinophils per hpf at the time of nasal brushing is also a limitation. This limitation was necessitated by our study design, as our goal was to validate the diagnostic accuracy and patient tolerance of a diagnostic test collected in clinic. Biopsy of sinonasal tissue in clinic was determined to be not feasible from the perspective of patient tolerance and willingness to participate in research. We attempted to address this limitation via two methods: (1) analysis of EPX levels in comparison to eosinophils per hpf in previously collected surgical pathology specimen, with a small subset of pre-operative patients having pathology results available shortly post-collection and (2) evaluating percent eosinophils on cell slides from cells collected via nasal cytology brush of the middle meatus. Furthermore, prior work by Smith et al. has validated the association between EPX activity levels in middle meatus nasal swabs and eosinophils per hpf in ethmoid biopsies taken simultaneously. Our study builds on this work, testing total EPX levels in nasal mucus which is likely to similarly correlate to eosinophils per hpf. Although this study evaluated total EPX levels in tissue homogenates, future work should compare total EPX levels in nasal mucus to EPX activity level in nasal mucus.
5 CONCLUSION
We demonstrate for the first time that in-clinic nasal brushing to collect nasal mucus can be utilized to measure EPX levels in nasal mucus. EPX levels correlate to clinical markers of type 2 inflammation and tissue eosinophilia and may provide a valuable diagnostic tool to delineate eosinophilic CRS. Moreover, EPX levels demonstrate a positive correlation with IL-5 levels. Further investigation is warranted to explore the dynamics of EPX levels in response to CRS treatments and the predictive capability of this biomarker regarding disease severity or treatment efficacy.