Mitochondrial DNA patterns in the Iberian Northern plateau: Population dynamics and substructure of the Zamora province
Abstract
Several studies have shown the importance of recent events in the configuration of the genetic landscape of a specific territory. In this context, due to the phenomena of repopulation and demographic fluctuations that took place in recent centuries, the Iberian Northern plateau is a very interesting case study. The main aim of this work is to check if recent population movements together with existing boundaries (geographical and administrative) have influenced the current genetic composition of the area. To accomplish this general purpose, mitochondrial DNA variations of 214 individuals from a population located in the Western region of the Iberian Northern plateau (the province of Zamora) were analyzed. Results showed a typical Western European mitochondrial DNA haplogroup composition. However, unexpected high frequencies of U5, HV0, and L haplogroups were found in some regions. The analyses of microdifferentiation showed that there are differences between regions, but no geographic substructure organization can be noticed. It can be stated that the differences observed in the genetic pool of the sampled area at regional level results from the mixture of different populations carrying new lineages into this area at different points in history. Am J Phys Anthropol 142:531–539, 2010. © 2010 Wiley-Liss, Inc.
Studies on European genetic diversity have focused mainly on large-scale variation and interpretations based on prehistoric events. However, the complex history of the Iberian Peninsula provides a suitable region to examine the demographic impact of recent historical events on the current genetic landscape. In this regard, Alvarez et al. (2009) and Adams et al. (2008), using Y chromosome data, proved how genetic diversity can be affected by recent population movements. Moreover, Larruga et al. (2001), using mitochondrial DNA (mtDNA) data, showed that recent time isolation, due to cultural differences, clearly influenced the genetic composition of Maragatos, a population located in the western mountainous region of the León province (Northern Spain).
Archaeological and historical data, available for the North-Western extreme of the Iberian plateau (see Fig. 1), reveals a temporal continuum for human activity since the first human settlements, dating from the mid-Pleistocene period (Santoja, 1992), to the present. One of the periods that critically affected the demographic history of the area spanned from the mid 8th century to the 11th century, when land along the Duero River line (Fig. 1a) remained practically uninhabited, becoming a defensive corridor between the Astur kingdom in the North and the Muslim kingdom in the South.
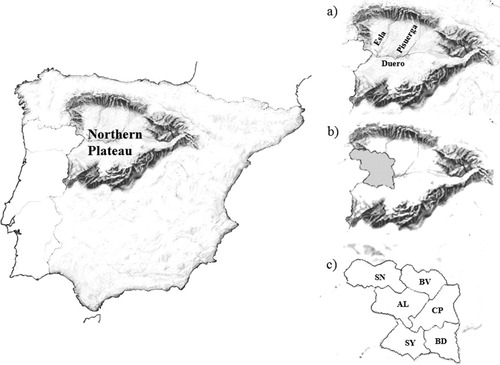
Geographical location of the Northern Plateau in the Iberian Peninsula. (a) Main rivers of the area; (b) position of the Zamora Province (gray color); (c) regional subdivision of the Zamora Province (codes for the regions as summarized in Table 1).
Concerning the Zamora province (Fig. 1b), historical data indicate that the territories gained by Northern kingdoms on the North of the Duero River at the end of the 8th century [present day Aliste, Benavente, Campos-Pan, and Sanabria regions (Fig. 1c)] were resettled by individuals from two main origins (Delibes and Moreta, 1995): (1) Mozarabs from the Southern Muslim kingdoms (town of Coria and cities of Toledo, Madrid, and Mérida); and (2) people from the North-Western kingdoms (present-day territories of Asturias, Galicia, and León). The southern areas of the Duero River [present-day Sayago and Bajo Duero regions (Fig. 1c)], were only definitively incorporated into the Astur kingdom in the 11th century, when resettlement of the area was promoted. Based on historical sources (Delibes and Moreta, 1995), three main locations have been proposed as the origins of the settlers: (1) North-Western Iberia, including people from the Northern regions of the Zamora province; (2) Al-Ándalus, mainly Mozarabs; and (3) Southern France, including individuals from areas known today as Gascony, Poitou, Périgord, Montpellier, Provence, and Lombardy.
As mentioned earlier, the North-Western extreme of the Iberian plateau, and within this the Zamora province, appears as a very interesting case-study population, which can be used to evaluate how relatively recent historical events could influence the genetic structure of human populations. Moreover, the geography, orography, and demography of the Zamora province also present interesting characteristics that could be used to test several additional hypotheses concerning the genetic substructure of the region.
To date, there are no genetic studies centered on the North-Western extreme of the Iberian plateau; thus, an integrative study of the region using different genetic systems is being undertaken. In this study, mtDNA variations of each of the six regions of the Zamora province are presented. The main goals of this work are (a) to infer if the different resettlements that took place during the period that ranged from the 8th to the 11th centuries are reflected in the current genetic composition of the area; (b) to check if the recent depopulation of the province (INE, 2008), mainly occurring in the Sanabria region during the 20th century, has had effects on the present-day genetic diversity; and (c) to infer the impact of natural and political boundaries on the genetic microdifferentiation of the province.
MATERIALS AND METHODS
Samples and comparative data sets
Two hundred and fourteen maternally unrelated individuals, born and with maternal origin in the Zamora province (confirmed up to the third generation) were sampled in regional health centers. The origin of the samples was defined taking into account the birthplace of the grandmother in one of the six regions in which the Zamora province can be subdivided, 32 samples from Aliste, 37 from Bajo Duero, 40 from Benavente, 41 from Campos-Pan, 31 from Sanabria, and 33 from Sayago. For all voluntary donors, appropriate informed consent and the birth places of all their known maternal ancestors up to the third generation were obtained under strictly confidential circumstances.
Total DNA from blood samples was extracted using the Jetquick Blood & Cell kit (Genomed) according to the manufacturer's specifications. For each sample, the mtDNA hypervariable region I (HVRI) was amplified and sequenced, between positions 16,024 and 16,399, and coding region informative polymorphisms (mtDNA positions: 1,018; 3,010; 4,580; 6,776; 7,028; 8,994; 9,055; 10,238; 11,251; 12,308; 12,705; 14,766) for haplogroup (Hg) assignment were analyzed by PCR-RFLP as previously detailed by Santos et al. (2003, 2004). Moreover, positions 10,400 and 10,873 were also analyzed; the fragment enclosing these mtDNA positions was amplified and sequenced using primers and conditions described by Ramos et al. (2009). Sequence reactions were carried out using the sequencing kit BigDye Terminator v.3 (Applied Biosystems) according to the manufacturer's specifications and run in an ABI 3130XL sequencer (Genomics Unit, Universitat Autònoma de Barcelona).
For comparative purposes 1,023 mtDNA HVRI Iberian sequences (nucleotide positions 16,024–16,383) were compiled and used for the diversity and genetic distance analysis (populations, sample size, and references are listed in Table 1). Additionally, 341 mtDNA HVRI sequences belonging to African macro-Hg L (nucleotide positions 16,090–16,365) were used for the shared haplotype (Ht) analysis of the African lineages. In this last dataset, original population data were grouped into the following geographic areas: Iberian Peninsula (Pereira et al., 2005); Northern Africa (Corte-Real et al., 1996; Rando et al., 1998; Krings et al., 1999; Brakez et al., 2001); South-eastern Africa (Pereira et al., 2001; Salas et al., 2002); Macaronesia (Rando et al., 1999; Brehm et al., 2002, 2003; Santos et al., 2003); and Central-West Africa (Watson et al., 1997; Rando et al., 1998; Plaza et al., 2004; Rosa et al., 2004).
| Population | Code | No. inhabitants | N | K (% K) | S (% S) | Ĥ ± sd | π ± sd | θK | θS |
|---|---|---|---|---|---|---|---|---|---|
| Zamoraa | ZA | 197,221 | 214 | 107 (50.00) | 80 (22.22) | 0.9510 ± 0.0112 | 0.01485 ± 0.00796 | 84.5001 | 13.4661 |
| Aliste | AL | 14,090 | 32 | 22 (68.75) | 35 (09.72) | 0.9577 ± 0.0235 | 0.01686 ± 0.00917 | 29.8144 | 8.6908 |
| Bajo Duero | BD | 21,688 | 37 | 24 (64.85) | 34 (09.44) | 0.9384 ± 0.0310 | 0.01313 ± 0.00730 | 28.4963 | 8.1446 |
| Benavente | BV | 40,462 | 40 | 29 (72.50) | 39 (10.83) | 0.9718 ± 0.0158 | 0.01219 ± 0.00683 | 45.8968 | 9.1688 |
| Campos-Pan | CP | 34,240 | 41 | 29 (70.73) | 40 (11.11) | 0.9659 ± 0.0183 | 0.01600 ± 0.00869 | 42.7824 | 9.3490 |
| Sanabria | SN | 10,283 | 31 | 20 (64.52) | 36 (10.00) | 0.8581 ± 0.0635 | 0.01210 ± 0.00684 | 23.2933 | 9.0113 |
| Sayago | SY | 9,786 | 33 | 22 (66.67) | 27 (07.50) | 0.9659 ± 0.0178 | 0.01825 ± 0.00984 | 27.6781 | 6.6527 |
| Mainland Spain | MS | 22,679,772 | 528 | 238 (45.08) | 124 (34.44) | 0.9463 ± 0.0079 | 0.01226 ± 0.00670 | 166.2373 | 18.1144 |
| Andalusiab,c | AN | 8,202,220 | 99 | 76 (76.77) | 74 (20.56) | 0.9734 ± 0.0114 | 0.01532 ± 0.00823 | 147.9744 | 14.3209 |
| Basque Countryd,e | BQ | 2,157,112 | 100 | 44 (44.00) | 41 (11.39) | 0.9287 ± 0.0186 | 0.00961 ± 0.00548 | 29.4604 | 7.9190 |
| Castileb | CA | 1,656,912 | 38 | 29 (76.32) | 38 (10.56) | 0.9644 ± 0.0213 | 0.01290 ± 0.00719 | 54.1794 | 9.0442 |
| Cataloniac | CT | 7,364,078 | 46 | 28 (58.33) | 35 (09.72) | 0.9179 ± 0.0345 | 0.01214 ± 0.00678 | 29.4099 | 7.9637 |
| Galiciaf,g | GA | 2,784,169 | 135 | 84 (62.22) | 75 (20.83) | 0.9544 ± 0.0138 | 0.01216 ± 0.00669 | 94.0145 | 13.6892 |
| Leonb | LE | 500,200 | 61 | 44 (72.13) | 53 (14.72) | 0.9410 ± 0.0256 | 0.01220 ± 0.00677 | 69.3584 | 11.3251 |
| Maragatosb | MG | 15,081 | 49 | 24 (48.98) | 34 (09.44) | 0.8954 ± 0.0362 | 0.01118 ± 0.00630 | 17.9520 | 7.6254 |
| North Portugalh | MNP | 3,294,006 | 187 | 114 (60.96) | 85 (23.61) | 0.9496 ± 0.0128 | 0.01499 ± 0.00803 | 122.9941 | 14.6409 |
| Braga | BR | 862,191 | 41 | 34 (82.93) | 43 (11.94) | 0.9793 ± 0.0150 | 0.01626 ± 0.00881 | 90.9577 | 10.0501 |
| Bragança | BG | 142,049 | 42 | 28 (66.67) | 36 (10.00) | 0.9489 ± 0.0246 | 0.01544 ± 0.00841 | 35.5456 | 8.3664 |
| Porto | PO | 1,820,752 | 67 | 44 (65.67) | 35 (16.39) | 0.9417 ± 0.0233 | 0.01467 ± 0.00796 | 54.6198 | 12.3575 |
| Viana do Castelo | VC | 251,676 | 17 | 14 (82.35) | 29 (08.06) | 0.9559 ± 0.0436 | 0.01668 ± 0.00936 | 34.6813 | 8.5780 |
| Vila Real | VL | 217,338 | 20 | 13 (65.00) | 22 (06.11) | 0.9105 ± 0.0538 | 0.00994 ± 0.00588 | 15.0011 | 6.2011 |
| Center Portugalh | MCP | 2,413,312 | 179 | 107 (59.78) | 87 (24.17) | 0.9610 ± 0.0107 | 0.01436 ± 0.00773 | 111.2688 | 15.0994 |
| South Portugalh | MSP | 4,419,562 | 183 | 108 (59.02) | 84 (23.33) | 0.9618 ± 0.0102 | 0.01662 ± 0.00881 | 109.7769 | 14.5229 |
- N, sample size; K, number of different haplotypes; S, number of polymorphic sites; Ĥ, gene diversity; π, nucleotide diversity; θK, theta estimator based on the number of different haplotypes; θS, theta estimator based on the number of polymorphic sites.
- a Present study.
- b Larruga et al., 2001.
- c Plaza et al., 2003.
- d Bertranpetit et al., 1995.
- e Alfonso-Sanchez et al., 2008.
- f Salas et al., 1998.
- g Gonzalez et al., 2003.
- h Pereira et al., 2004.
Data analysis
Samples were assigned to Hgs using the combined information of HVRI and coding region variations following the updated phylogenetic classification proposed by van Oven and Kayser (2009).
The Hg frequencies were determined by counting and the Bayesian 0.95 credible region (95% CR) was calculated using the SAMPLING programme (Macaulay, personal communication). The Hg distribution observed in the six regions of Zamora was compared using the exact test of population differentiation, as described in Raymond and Rousset (1995). To assess global differences in the Hg composition of a specific area, correspondence analysis, using Hgs and regions, and χ2 tests were conducted in the SPSS ver. 15.0.1 software (SPSS Inc.).
Because the impact of genetic drift on the distribution of mtDNA Hts in a population is not equally captured by different diversity indicators, HVRI sequences between positions 16,024 and 16,383 were used to estimate several standard and molecular diversity indices using Arlequin 3.1 software (Excoffier et al., 2005), namely, the Gene diversity (Ĥ) (Nei, 1987), the number of different Ht (K), the number of polymorphic sites (S), the nucleotide diversity (π) (Tajima, 1983; Nei, 1987), and the theta estimators based on the number of polymorphic sites (θS) (Watterson, 1975) and on the number of different Hts (θk) (Ewens, 1972), this last estimator being more sensitive to genetic drift according to Helgason et al. (2003). The Tamura and Nei (1993) nucleotide substitution model with a gamma correction (a = 0.205) (Bandelt et al., 2006) was used.
Inferences regarding past demographic effects on the genetic variation in current populations were conducted by comparing mismatch distributions of pairwise nucleotide differences between Hts to those expected under a sudden population expansion model (Slatkin and Hudson, 1991; Rogers and Harpending, 1992; Rogers, 1995). Statistical significance for the mismatch distributions was obtained using a goodness-of-fit test based on thesum of squared deviations between the observed and expected distributions (Schneider and Excoffier, 1999) and the Harpending's raggedness index (rg) (Harpending, 1994), after 1,000 simulations, using the estimated parameters of the expected distribution for a population expansion. Demographic history was further assessed using the Tajima's D statistic (Tajima, 1989).
To investigate the relation between populations and the effect of natural boundaries on the genetic structure, AMOVA analyses (Excoffier et al., 1992), using both Hg frequencies and HVRI sequence variation, were conducted, with the Arlequin 3.1 software (Excoffier et al., 2005), to check all possible based geographical structures, namely: (1) all the regions within one group; (2) regions on the north of the Duero River versus southern regions; (3) Eastern regions versus western regions; and (4) regions divided by the Duero and Esla rivers.
To analyze the situation of the Zamora regions in the context of the adjacent Iberia populations (Galicia, León, and Bragança), and although the sampling unities are different, Slatkin's linearized FST pairwise genetic distance (Slatkin, 1995) was calculated, using HVRI sequences, with the Arlequin 3.1 (Excoffier et al., 2005). The matrices obtained were then represented in two-dimensional space by means of the multidimensional scaling (MDS) procedure using the SPSS ver. 15.0.1 (SPSS Inc.). Moreover, Barrier software vs. 2.2, which implements the Monmonier algorithm (Manni et al., 2004), was used to calculate barriers relating to geography and the Slatkin's linearized FST pairwise genetic distance (Slatkin, 1995). To assess the robustness of calculated barriers, 100 resampled bootstrap data sets were generated using SAS software (SAS Institute Inc.) [Calculation available on request].
RESULTS
Hg and HVRI variation
Detailed mtDNA results are presented as online supporting material (Supporting Information Table 1); the 214 sampled individuals represent 123 distinct Ht.
Frequencies of mtDNA Hg found in the Zamora province and in each region are listed in Table 2. The Hg distribution in the whole province showed, in general, typical frequencies for a Western European population; however, unexpected high frequencies of haplogroups (Hgs) HV0 and L were found in the Bajo Duero and Sayago regions (χ2 test, P = 0.001 and P < 0.000, respectively), and a high frequency of Hg U5 was found in the Aliste region (χ2 test, P = 0,003) when compared with the rest of regions.
| AL (N = 32) | BD (N = 37) | BV (N = 40) | CP (N = 41) | SN (N = 31) | SY (N = 33) | Total ZA (N = 214) | |
|---|---|---|---|---|---|---|---|
| L1b | 0.0 (0.1−10.6) | 0.0 (0.1−9.3) | 0.0 (0.1−7.0) | 0.0 (0.1−8.4) | 3.2 (0.8−16.2) | 9.1 (3.3−23.7) | 1.9 (0.8−4.7) |
| L2b | 0.0 (0.1−10.6) | 5.4 (1.7−17.7) | 0.0 (0.1−7.0) | 0.0 (0.1−8.4) | 0.0 (0.1−10.9) | 0.0 (0.1−10.3) | 0.9 (0.3−3.3) |
| L3b | 0.0 (0.1−10.6) | 2.7 (0.6−13.8) | 0.0 (0.1−7.0) | 0.0 (0.1−8.4) | 0.0 (0.1−10.9) | 9.1 (3.3−23.7) | 1.9 (0.8−4.7) |
| M1 | 0.0 (0.1−10.6) | 0.0 (0.1−9.3) | 0.0 (0.1−7.0) | 2.4 (0.6−12.6) | 0.0 (0.1−10.9) | 0.0 (0.1−10.3) | 0.5 (0.1−2.6) |
| N1b | 3.1 (0.7−15.8) | 0.0 (0.1−9.3) | 0.0 (0.1−7.0) | 2.4 (0.6−12.6) | 0.0 (0.1−10.9) | 0.0 (0.1−10.3) | 0.9 (0.3−3.3) |
| I | 0.0 (0.1−10.6) | 0.0 (0.1−9.3) | 0.0 (0.1−7.0) | 4.9 (1.5−16.2) | 3.2 (0.8−16.2) | 0.0 (0.1−10.3) | 1.4 (0.5−4.0) |
| W | 0.0 (0.1–10.6) | 2.7 (0.6–13.8) | 0.0 (0.1–7.0) | 2.4 (0.6–12.6) | 0.0 (0.1–10.9) | 0.0 (0.1–10.3) | 0.9 (0.3–3.3) |
| X | 0.0 (0.1–10.6) | 2.7 (0.6–13.8) | 0.0 (0.1–7.0) | 0.0 (0.1–8.4) | 0.0 (0.1–10.9) | 3.0 (0.7–15.3) | 0.9 (0.3–3.3) |
| R0 | 0.0 (0.1–10.6) | 0.0 (0.1–9.3) | 0.0 (0.1–7.0) | 0.0 (0.1–8.4) | 3.2 (0.8–16.2) | 0.0 (0.1–10.3) | 0.5 (0.1–2.6) |
| R0a | 0.0 (0.1–10.6) | 0.0 (0.1–9.3) | 0.0 (0.1–7.0) | 2.4 (0.6–12.6) | 0.0 (0.1–10.9) | 0.0 (0.1–10.3) | 0.5 (0.1–2.6) |
| R1 | 3.1 (0.7–15.8) | 0.0 (0.1–9.3) | 0.0 (0.1–7.0) | 0.0 (0.1–8.4) | 0.0 (0.1–10.9) | 0.0 (0.1–10.3) | 0.5 (0.1–2.6) |
| HV0 | 3.1 (0.7–15.8) | 8.1 (2.9–21.4) | 0.0 (0.1–7.0) | 0.0 (0.1–8.4) | 3.2 (0.8–16.2) | 15.2 (6.8–31.1) | 4.7 (2.6–8.4) |
| HV | 3.1 (0.7–15.8) | 0.0 (0.1–9.3) | 0.0 (0.1–7.0) | 0.0 (0.1–8.4) | 3.2 (0.8–16.2) | 0.0 (0.1–10.3) | 0.9 (0.3–3.3) |
| V | 3.1 (0.7–15.8) | 8.1 (2.9–21.4) | 5.0 (1.5–16.5) | 2.4 (0.6–12.6) | 3.2 (0.8–16.2) | 3.0 (0.7–15.3) | 4.2 (2.3–7.8) |
| H* | 3.1 (0.7–15.8) | 5.4 (1.7–17.7) | 30.0 (18.1–45.5) | 7.3 (2.7–19.5) | 16.1 (7.2–32.8) | 9.1 (3.3–23.7) | 12.1 (8.4–17.2) |
| H1 | 18.8 (9.0–35.5) | 18.9 (9.6–34.3) | 15.0 (7.2–29.2) | 24.4 (13.9–39.5) | 22.6 (11.5–40.0) | 3.0 (0.7–15.3) | 17.3 (12.8–22.9) |
| H3 | 9.4 (3.4–24.3) | 10.8 (4.4–24.8) | 10.0 (4.1–23.1) | 9.8 (4.0–22.6) | 3.2 (0.8–16.2) | 15.2 (6.8–31.1) | 9.8 (6.5–14.5) |
| J* | 12.5 (5.1–28.2) | 8.1 (2.9–21.4) | 2.5 (0.6–12.9) | 4.9 (1.5–16.2) | 9.7 (3.5–25.0) | 3.0 (0.7–15.3) | 6.5 (4.0–10.7) |
| J1 | 0.0 (0.1–10.6) | 2.7 (0.6–13.8) | 0.0 (0.1–7.0) | 0.0 (0.1–8.4) | 0.0 (0.1–10.9) | 0.0 (0.1–10.3) | 0.5 (0.1–2.6) |
| J1b1 | 0.0 (0.1–10.6) | 0.0 (0.1–9.3) | 0.0 (0.1–7.0) | 4.9 (1.5–16.2) | 0.0 (0.1–10.9) | 0.0 (0.1–10.3) | 0.9 (0.3–3.3) |
| J2a1 | 0.0 (0.1–10.6) | 0.0 (0.1–9.3) | 5.0 (1.5–16.5) | 0.0 (0.1–8.4) | 0.0 (0.1–10.9) | 0.0 (0.1–10.3) | 0.9 (0.3–3.3) |
| J2b | 0.0 (0.1–10.6) | 0.0 (0.1–9.3) | 0.0 (0.1–7.0) | 7.3 (2.7–19.5) | 0.0 (0.1–10.9) | 9.1 (3.3–23.7) | 2.8 (1.3–6.0) |
| T* | 0.0 (0.1–10.6) | 2.7 (0.6–13.8) | 0.0 (0.1–7.0) | 0.0 (0.1–8.4) | 0.0 (0.1–10.9) | 0.0 (0.1–10.3) | 0.5 (0.1–2.6) |
| T1 | 3.1 (0.7–15.8) | 0.0 (0.1–9.3) | 2.5 (0.6–12.9) | 0.0 (0.1–8.4) | 0.0 (0.1–10.9) | 0.0 (0.1–10.3) | 0.9 (0.3–3.3) |
| T2b | 6.3 (1.9–20.2) | 0.0 (0.1–9.3) | 0.0 (0.1–7.0) | 2.4 (0.6–12.6) | 3.2 (0.8–16.2) | 0.0 (0.1–10.3) | 1.9 (0.8–4.7) |
| T2c | 0.0 (0.1–10.6) | 0.0 (0.1–9.3) | 0.0 (0.1–7.0) | 0.0 (0.1–8.4) | 3.2 (0.8–16.2) | 0.0 (0.1–10.3) | 0.5 (0.1–2.6) |
| T2e | 0.0 (0.1–10.6) | 2.7 (0.6–13.8) | 0.0 (0.1–7.0) | 0.0 (0.1–8.4) | 0.0 (0.1–10.9) | 0.0 (0.1–10.3) | 0.5 (0.1–2.6) |
| U* | 0.0 (0.1–10.6) | 0.0 (0.1–9.3) | 7.5 (2.7–19.9) | 2.4 (0.6–12.6) | 6.5 (2.0–20.8) | 0.0 (0.1–10.3) | 2.8 (1.3–6.0) |
| U2 | 0.0 (0.1–10.6) | 0.0 (0.1–9.3) | 2.5 (0.6–12.9) | 4.9 (1.5–16.2) | 0.0 (0.1–10.9) | 0.0 (0.1–10.3) | 1.4 (0.5–4.0) |
| U5 | 12.5 (5.1–28.2) | 2.7 (0.6–13.8) | 7.5 (2.7–19.9) | 7.3 (2.7–19.5) | 3.2 (0.8–16.2) | 6.1 (1.9–19.7) | 6.5 (4.0–10.7) |
| U5a | 6.3 (1.9–20.2) | 0.0 (0.1–9.3) | 2.5 (0.6–12.9) | 2.4 (0.6–12.6) | 3.2 (0.8–16.2) | 0.0 (0.1–10.3) | 2.3 (1.0–5.3) |
| U5a1 | 3.1 (0.7–15.8) | 8.1 (2.9–21.4) | 2.5 (0.6–12.9) | 2.4 (0.6–12.6) | 0.0 (0.1–10.9) | 6.1 (1.9–19.7) | 3.7 (1.9–7.2) |
| U5b | 9.4 (3.4–24.3) | 0.0 (0.1–9.3) | 2.5 (0.6–12.9) | 0.0 (0.1–8.4) | 0.0 (0.1–10.9) | 0.0 (0.1–10.3) | 1.9 (0.8–4.7) |
| U6a1a | 0.0 (0.1–10.6) | 0.0 (0.1–9.3) | 0.0 (0.1–7.0) | 0.0 (0.1–8.4) | 3.2 (0.8–16.2) | 0.0 (0.1–10.3) | 0.5 (0.1–2.6 |
| K | 0.0 (0.1–10.6) | 8.1 (2.9–21.4) | 5.0 (1.5–16.5) | 2.4 (0.6–12.6) | 6.5 (2.0–20.8) | 9.1 (3.3–23.7) | 5.1 (2.9–9.0) |
As regards the HVRI Hts, Sanabria presents the lowest percentage of distinct Hts (64.52%), followed by Bajo Duero (64.85%) and Sayago (66.67) (Table 1). Moreover, the analysis of shared Hts proves that all the regions present low percentages of shared Hts with the remaining regions (Table 3). Nevertheless, the percentage of shared Hts is higher between adjacent regions with the exception of those which have the Duero River as a border: Campos-Pan and Bajo Duero; Campos-Pan and Sayago; and Aliste and Sayago. In Sanabria, the percentage of private lineages or nonshared Hts between populations (diagonal values in Table 3), revealed low levels, because it shares Ht, mainly the Hg founder motifs, with the rest of the regions.
| Shared Ht (%) | ||||||
|---|---|---|---|---|---|---|
| AL | BD | BV | CP | SN | SY | |
| AL | 54.55 | 12.50 | 17.24 | 13.79 | 23.81 | 09.01 |
| BD | 13.64 | 66.67 | 10.34 | 10.34 | 19.05 | 27.27 |
| BV | 22.73 | 12.50 | 58.62 | 17.24 | 33.33 | 18.18 |
| CP | 18.18 | 12.50 | 17.24 | 58.62 | 28.57 | 27.27 |
| SN | 22.73 | 16.67 | 24.14 | 20.79 | 33.33 | 22.72 |
| SY | 09.01 | 25.00 | 13.79 | 10.34 | 23.81 | 59.09 |
- The percentages in each column were calculated using number of Ht in the regions that represent the column title; each percentage represents the proportion of Ht from the column population that are also present in the row population. Diagonal values represent the percentage of private lineages or non shared Ht between populations (marked in bold).
Diversity indicator values, based on the HVRI sequences, found in regions of the Zamora province and in other Iberian populations are summarized in Table 1. The results for the Zamora province show similar values of gene diversity and nucleotide diversity as the mean of mainland Spain and Portugal, however, the value of θK is lower. In the analyzed regions, Sanabria and Bajo Duero showed small values of gene diversity and also of θK. The gene diversity in Sanabria is in fact the lowest of the Iberian populations used for comparison. Also, mismatch distribution analyses and Tajima neutrality tests (Table 4) point respectively to unimodal patterns of mismatch distributions and significant negative D values.
| Population | Tajima's D | SSD | Raggedness index |
|---|---|---|---|
| Zamora | −2.013* | 0.00029 | 0.00704 |
| Aliste | −1.482 | 0.00341 | 0.01126 |
| Bajo Duero | −1.735* | 0.00180 | 0.01256 |
| Benavente | −2.031* | 0.00338 | 0.02634 |
| Campos-Pan | −1.638* | 0.00062 | 0.00706 |
| Sanabria | −2.126* | 0.00921 | 0.02926 |
| Sayago | −0.683 | 0.00178 | 0.01006 |
| Mainland Spain | −2.298** | 0.00046 | 0.00952 |
| North Portugal | −2.106* | 0.00098 | 0.00594 |
| Centre Portugal | −2.186* | 0.00042 | 0.00714 |
| South Portugal | −2.018* | 0.00052 | 0.00650 |
- SSD, sum of square deviations between the observed and the expected mismatch distribution.
- * P < 0.05.
- ** P < 0.001.
Population substructure
To investigate the genetic microdifferentiation in the Zamora province, an exhaustive analysis, using different methodologies, was implemented.
The Hg distribution of pairs of regions was compared, and the results of the exact test of population differentiation are presented in Supporting Information Table 2 (online supporting data). After the Bonferroni correction for multiple tests, Sayago showed significant differences compared to Aliste and Benavente, and no differences were found between the other regions. Moreover, Hg and HVRI-based AMOVA revealed that there is significant variation among the six regions (HVRI: FST = 0.01239, P = 0.0029; Hg: FST = 0.01376, P = 0.0059).
To better understand the Hg distribution on a microgeographic level, a correspondence analysis based on the Hg distribution was conducted (see Fig. 2). The European Hg U5 (Macaulay et al., 1999) and its sub-Hgs are well represented in all regions with an extremely high frequency in Aliste (31.25%). Hg HV0 (or pre*V) (Torroni et al., 2001) appears, in the positive portion of dimension 1, associated with the southern regions of Bajo Duero (8.1%) and Sayago (15.2%). Apart from the high frequency of HV0, Bajo Duero and Sayago also present high incidences of sub-Saharan lineages that account for 18.2% of the individuals in Sayago (L1b and L3) and 5.4% in Bajo Duero (L2b).
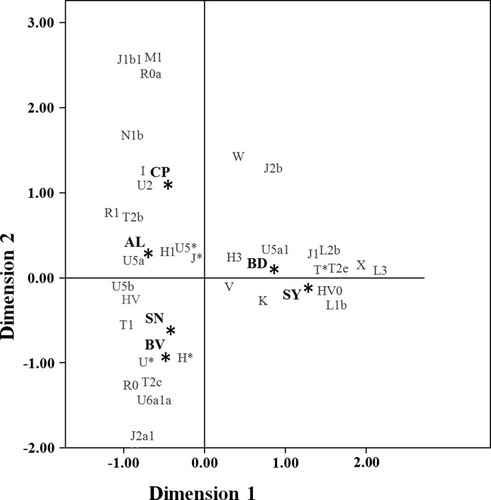
Correspondence analysis based on the haplogroup distribution on each of the six regions of Zamora.
It seems clear, from all the analysis, that Sayago represents a genetic outlier in the Zamora province. Nevertheless, if more attention is placed on correspondence analysis (see Fig. 2), a geographic structure determined mainly by the Duero River could be inferred. However, in the AMOVA analysis of different population groups, using both HVRI and Hg data, no significant differences were detected between the defined areas, and significant differences were found among populations within groups.
The MDS representation of Slatkin's linearized FST pairwise genetic distance (Slatkin, 1995), based on HVRI sequences between the Zamora regions and the adjacent Iberian populations (see Fig. 3), reveals that the majority of the analyzed regions in Zamora are located around the center of axis showing a great homogeneity. However, the Sayago and Aliste regions appear far from the principal population group and there is also a great distance between them. As Sayago and Aliste (and also Sanabria) have a border with the Portuguese region of Bragança, great affinities with this Portuguese region could a priori be expected; however, only Aliste appears near Bragança. Barrier analysis (Manni et al., 2004) was performed to find out more about the geographic and/or political boundaries that could affect the genetic distances observed (see Fig. 4). The analysis of the Zamora regions and the adjacent sampled areas showed three principal barriers in the zone: the first one involves the Sayago region; in accordance to previous results, it represents the stronger barrier based on genetic and geographic distances; the second one separates Bragança from all of the Spanish areas (the areas of Zamora and Galicia), which shows the importance of the Spanish-Portuguese border as a barrier to gene flow between adjacent territories; and the third barrier appears around the Aliste region, isolating it from the rest of the regions.
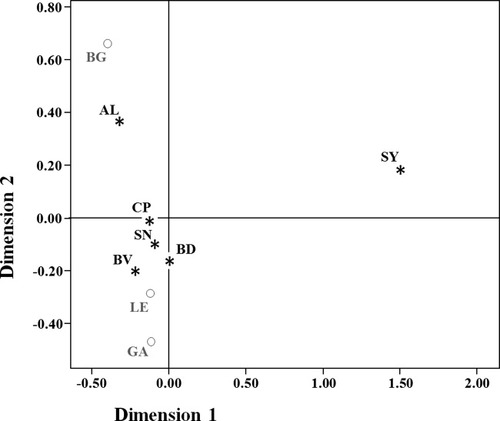
Multidimensional scaling representation of the Slatkin genetic distances, based on HVRI sequences.
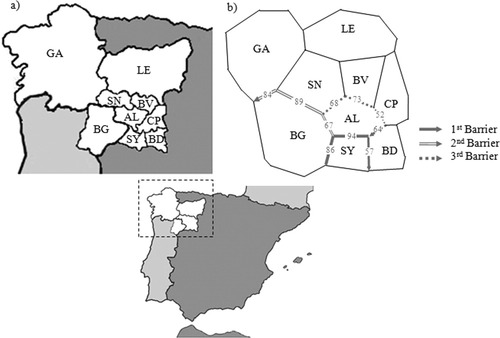
(a) Situation of the Zamora province and the adjacent areas in the North West portion of the Iberian Peninsula. (b) Three main barriers detected in the Barrier analysis based on Monmonier algorithm elaborates from Stalking genetic distances on 100 bootstrap matrices obtained by randomly resampling the original data. The thickness of each edge of a barrier is proportional to the number of times it was included in the computed barriers (numbers on edges).
African influence
In the province of Zamora, if the total number of African lineages are taken into account (Hgs L1b, L2b, L3, M1, and U6a1a), the contribution represents 5.7% of the total Hg composition. As regards North African lineages, only one mtDNA type belonging to U6a1a Hg was found in the province, more precisely in the Sanabria region. The Ht ZA117 (16172-16183C-16189-16219-16239-16278) is observed in Algeria, Italy, and, inside the Iberian Peninsula, it is found in the North-Western populations of León and Cantabria (Larruga et al., 2001; Maca-Meyer et al., 2003; Pereira et al., 2005; van Oven and Kayser, 2009). There is a low frequency of U6 Hg in Iberia, its presence being higher in the North: 5.35% in Northern Portugal, 2.17% in Galicia, and 8.1% in Maragatos (León) (Salas et al., 1998; Larruga et al., 2001; Pereira et al., 2005). Hg M1 is found in Andalusia and Central Portugal although the Ht ZA6 (16093-16129-16148-16183C-16189-16249-16311), found in Campos-Pan, does not correspond to any of the previously detected Ht in Iberia.
As regards sub-Saharan Hgs (L1b, L2b, and L3b), the high frequency found in the southern regions of Zamora, 18.2% in Sayago and 8.1% in Bajo Duero, is comparable to that described for the South of Portugal, but it does not have any parallels with any other analyzed areas in the Northern part of Iberia (Pereira et al., 2005).
To try to shed some light on the precedence of sub-Saharan Ht in the Iberian Northern plateau, a shared Ht analysis was conducted. Four individuals typed as L1b, representing two different Ht, were found in the province (three in Sayago and one in Sanabria). Ht ZA1 (16126-16187-16189-16223-16264-16270-16278-16293-16311) is shared mainly with Central-West Africa, North Africa, and Macaronesia. Ht ZA2 (16126-16187-16189-16223-16264-16270-16278-16293-16311-16362) is shared with Macaronesia and Iberian samples. The L2b Ht found in the province corresponds to two individuals from the Bajo Duero region, Ht ZA3 (16114A-16129-16213-16223-16278) shared motif with samples from Central-West Africa, Macaronesia, the Iberian Peninsula, and Northern Africa. The two L3b Ht are present in four individuals (one in Bajo Duero and three in Sayago). These share the motif Ht ZA4 (16223-16278-16362) with Central-West Africa and the Iberian Peninsula. On the contrary, Ht ZA5 (16209-16223-16278-16362) is not found in any of the populations considered and differs from the Ht ZA4 by one mutation step. Thus, this Ht could have derived from ZA4 in the peninsula by a new mutation.
DISCUSSION
The mtDNA composition and diversity observed in the Zamora province shows perfect affinity with other Iberian populations. However, some differences between regions are detected.
Concerning the mtDNA diversity observed, the Sanabria region presents the lowest values when compared with other Iberian populations. Because Sanabria has one of the lowest numbers of inhabitants (Table 1), it may be hypothesized that the low diversity observed in this region could be related to the number of inhabitants. Taking into account that, in small populations, drift will be the primary evolutionary force acting, particularly reducing the number of rare alleles (Helgason et al., 2003) and, that the number of inhabitants is related with θK (r = 0.702; P < 0.001), it seems that the differences in the diversity observed in the regions of Zamora are related to population size. Moreover, both Tajima's D and mismatch distribution analysis reveal quite similar demographic histories for the different regions of the province, and it appears that the reduction by ∼70% of Sanabria inhabitants in the last 50 years has not had a detectable impact on the genetic landscape of this region. However, this result must be interpreted with caution, because recent changes in population size may not be detectable in mismatch distribution analyses due to threshold effects, time lags, or earlier demographic events that may mask the effects of recent events (Rogers and Harpending, 1992).
The analyses of genetic microdifferentiation showed that there are differences between regions, both at Hg and HVRI levels, but no geographic substructure organization (determined by rivers) can be noticed, because the observed differences appeared at the regional level mainly in Sayago and Aliste. Moreover, the diversity values obtained in these two regions and the observed intra Hg diversity are not consistent with a profile of isolated populations that could be used as a justification for the observed isolation. Barrier analysis is particularly informative about the differentiation of these two regions (see Fig. 4). Sayago appears as the most differentiated population with respect to both the province and the Portuguese region of Bragança. However, this first barrier presents its thinner portion with the Bajo Duero region due to the high presence of African lineages in both regions in contrast to the rest of the Province. As regards Aliste, this region appears to present some affinity with the Portuguese region of Bragança. The border between those populations is thinner when compared with the other regions that reveal population affinities. However, according to barrier analysis, the political frontier along the entire Portuguese border with Zamora acts as a strong boundary to gene flow, isolating all the Spanish areas from the Portuguese area of Bragança. This point, however, must be further researched with appropriate sample representation, particularly for the Portuguese region of Bragança.
One of the distinctions between the mtDNA composition of the Iberian Peninsula with respect to other European populations is the presence of North African and sub-Saharan lineages [for revision, see Arroyo-Pardo et al. (2007)]. In Zamora, both North African and sub-Saharan mtDNA lineages were found. It has been suggested that U6 and M1 Hgs, detected in low frequencies in Zamora, have been involved in the dispersal of Upper Palaeolithic Levantine people to North Africa along the south Mediterranean coastal areas (Olivieri et al., 2006). In this scenario, prehistoric links between North Africa and Iberia could explain the presence of this Hg in the Northern part of Iberia. The identification of a M1 mtDNA African lineage in a Basque necropolis dating back to the 6th–7th centuries (Izagirre et al., 2005) together with cattle from the Bronze Age (Anderung et al., 2005) with mtDNA African lineage support this hypothesis. However, paying heed to the low diversity of these Hg in Iberia, a more recent North African contribution that claims that it may be due to the flexible procreation between Christians and Muslims (females) (Pereira et al., 2005) is plausible. Another explanation is the relocation of moriscos, a hypothesis recently proposed by Adams et al. (2008) based on Y chromosome data and supported by historical data available for the studied region (Martin, 2003).
As regards sub-Saharan lineages, it is well known that during the 16th–19th centuries, African slaves were captured along the West African Coast and were frequently transported to Cape Verde (Macaronesia region). This archipelago served, from the beginning of the slave trade, as a kind of platform that connected the African continent to Europe, America, and India from which slaves were transported to different regions, including the other Macaronesia archipelagos (Canary, Madeira, and Azores) and mainland Portugal (Comissão Nacional para as Comemorações dos Descobrimentos Portugueses, 1999). Thus, paying heed to the sharing Ht analyses, it seems that the slave trade, during the 16th–19th centuries, better explains the African sub-Saharan lineages found in the Iberian Peninsula (including those found in the Zamora province). However, in contrast to the well- documented presence of slaves in the Portuguese territories (Comissão Nacional para as Comemorações dos Descobrimentos Portugueses, 1999), the same evidence does not exist for mainland Spain. In the Zamora province, there is only one reference to the presence of slaves in the province (Carbajo Martin, 1995). As the Hts found in the area are also shared with North African populations, we cannot discard the possibility that these lineages derived from the North African Muslim permanence in the Iberian Peninsula. A great number of Berber troops relocated their family groups to the gained territories (Salvatierra and Canto, 2008). Thus, this phenomenon could explain the presence of sub-Saharan lineages.
Finally, it is important to emphasize the importance of regional studies, which can reveal results that would be obscured in an overall view of a major area. However, it is worth noting that this kind of strategy could be limited by sample size requirements.




