Mutational spectrum of dystrophinopathies in Singapore: Insights for genetic diagnosis and precision therapy
Funding information: Biomedical Research Council of Singapore, Grant/Award Number: 06-1-21-19476; National Medical Research Council, Grant/Award Number: 091212/2004; 1056/2011
Abstract
Duchenne and Becker muscular dystrophies (DMD/BMD) are X-linked recessive disorders caused by mutations in the DMD gene. Emerging therapies targeting patients with specific mutations are now becoming a reality for many of these patients. Precise molecular diagnosis is essential to facilitate the identification of possible new treatments for patients in the local context. In this study, we screened 145 dystrophinopathic patients in Singapore and assessed their molecular status for eligibility to current emerging genetic therapies. Overall, 140 (96.5%) of all patients harbored pathogenic DMD mutations comprising 95 exonic deletions (65.5%), 14 exonic duplications (9.7%), and 31 pathogenic small mutations (21.4%). Nonsense and frameshift mutations constitute 83.9% of all the small mutations. We found 71% (103/145) of all Singaporean dystrophinopathy patients to be theoretically amenable for exon skipping, either through skipping of single (53.1%) or multiple exons (17.9%). This approach is applicable to 81.1% (77/95) of patients carrying deletions and 83.9% (26/31) of those with small mutations. Eteplirsen induced skipping of exon 51 is applicable to 12.4% of local patients. Nonsense read-through therapy was found to be applicable in another 12.4% of all patients. Mutation screening is crucial for providing insights into the underlying genetic signature of the disease in the local population and contributes toward existing information on DMD mutations in Asia and globally. This will guide future targeted drug development and clinical trial planning for this disease.
1 INTRODUCTION
Dystrophinopathies are X-linked recessive neuromuscular disorders caused by mutations in the DMD gene [MIM #300377] and cover a spectrum of severe to mild phenotypes including Duchenne muscular dystrophy (DMD), Becker muscular dystrophy (BMD) and X-linked dilated cardiomyopathy (DCM). DMD (MIM #310200) is the most common fatal genetic disorder with a worldwide incidence of 1 in 3,500 live male births (Emery, 2002), while BMD (MIM #300376) affects about 1 in 18,450 male births (Bushby, Thambyayah, & Gardner-Medwin, 1991). DMD is characterized by loss of ambulation by around 12 years, progressive muscle degeneration and death by around the second or third decade of life due to left ventricular dysfunction or respiratory failure (Worton & Thompson, 1988). In contrast, BMD patients have milder disease manifestations and are diagnosed at a later age, often remain ambulant until later in life and generally have near normal life expectancies, although more severe cases have been noted (Aartsma-Rus, Janson, van Ommen, & van Deutekom, 2007). Both DMD and BMD are allelic disorders caused by different types of mutations in the DMD gene. In DMD, out-of-frame or nonsense mutations lead to truncated nonfunctional dystrophin protein, while the milder BMD phenotype is associated with mutations that maintain the open reading frame resulting in the production of semi-functional protein (Monaco, Bertelson, Liechti-Gallati, Moser, & Kunkel, 1988). Hence, molecular diagnosis is critical in DMD/BMD patients as it delineates the underlying genotype to guide disease management and prognosis.
The majority of the causative mutations resulting in DMD/BMD phenotype are large deletions or duplications, accounting for 65 and 5–15% of cases, respectively and the remaining fraction of pathogenic variations are known to be caused by small mutations or rearrangements (Hu, Ray, Murphy, Thompson, & Worton, 1990; Muntoni, Torelli, & Ferlini, 2003; Roberts, Bobrow, & Bentley, 1992). Deletions and duplications spanning one or more exons are detected using routine diagnostic methods such as multiplex PCR or multiplex ligation dependent probe amplification (MLPA) (Abbs, Yau, Clark, Mathew, & Bobrow, 1991; Beggs, Koenig, Boyce, & Kunkel, 1990; den Dunnen & Beggs, 2006; Schouten et al., 2002; Schwartz & Dunø, 2004; White et al., 2002). Methods used to detect more subtle sequence variations include mRNA transcript analysis, protein truncation test, denaturing gradient gel electrophoresis, single condition amplification/internal primer sequencing, single-strand conformational polymorphism analysis, nanoparticle assays, and so forth (Flanigan et al., 2003; Gardner, Bobrow, & Roberts, 1995; Hofstra et al., 2004; Low et al., 1996; Qin, Yim, Lai, & Yung, 2010; Roest et al., 1993; Tay, Hwee Hoon Khng, Poh Sim Low, & Poh San Lai, 2006; Tuffery et al., 1995; Tuffery et al., 1998; Tuffery, Moine, Demaille, & Claustres, 1993). Linkage markers were previously used for carrier screening in DMD but found to be useful only in families that were informative and who did not carry intragenic gene recombination events (Giliberto et al., 2003; Lai, Chiu, Low, Lee, & Tay, 1995; Schwartz, Tarleton, Popovich, Seltzer, & Hoffman, 1992). Use of such markers has since been replaced by other techniques such as Sanger sequencing which can detect both patients and at-risk family members carrying small sequence mutations. Additionally, next generation sequencing-based tests can detect both deletion or duplication mutations in addition to the small sequence mutations (Evilä, Arumilli, Udd, & Hackman, 2016; Gonorazky et al., 2016; Hegde et al., 2008; Okubo et al., 2016; Roberts, Barby, Manners, Bobrow, & Bentley, 1991; Wang et al., 2014; Wei et al., 2014; Wu, Brady, Shoffner, & Tarnopolsky, 2018).
There is no cure for DMD and current treatment involves supportive management and control of symptoms. There have been a growing number of molecular therapeutic approaches for DMD with some in clinical trials or under development. These include strategies such as adeno-associated virus (AAV) mediated delivery of mini-dystrophin, gene editing using Crispr-Cas9, exon skipping induction, and suppression of premature stop codons (Chamberlain & Chamberlain, 2017; Finkel, 2010; Takeshima et al., 2001; van Deutekom et al., 2001; Xu et al., 2016; Zhang & Duan, 2012). Among some of the most promising approaches are those targeted toward specific mutations. In the exon skipping strategy, antisense oligonucleotides (AONs) targeted to specific sequences mediate the exclusion of out-of-frame exon(s) in the transcript to convert out-of-frame mutations to in-frame mutations leading to a milder phenotype for DMD patients (Aartsma-Rus et al., 2009; Aartsma-Rus et al., 2017; Kinali et al., 2009; Nakamura, 2017; Pramono et al., 2012; Shimizu-Motohashi, Miyatake, Komaki, Takeda, & Aoki, 2016; Takeshima et al., 2006; Wee et al., 2008; Wilton et al., 2007). The recent breakthrough in exon skipping therapeutics came with the release of the first and the only FDA approved drug, Eteplirsen (Exondys 51) which is applicable for DMD patients amenable for exon 51 skipping (https://www.fda.gov/newsevents/newsroom/pressannouncements/ucm521263.htm). To allow for the treatment of additional groups of patients, AONs targeting other exons must be developed and marketed, and currently, clinical trials with AONs targeting exon 53, Golodirsen (SRP-4053), and exon 45, NS-065/NCNP-01 (NS Pharma, Paramus, NJ) are underway (Nguyen & Yokota, 2017). For the other therapeutic strategy targeting ribosomal read-through of premature stop codons, Ataluren (Translarna; PTC Therapeutics USA) received conditional approval in Europe for DMD patients with nonsense mutations (http://ir.ptcbio.com/news-releases/news-release-details/ptc-therapeutics-receives-conditional-approval-european-union).
Hence, precise molecular analysis of the underlying mutations in patients is required for accurate application of the appropriate therapeutic strategy (Aartsma-Rus, Ginjaar, & Bushby, 2016). Here we report the largest and first ever study in 145 dystrophinopathic patients diagnosed in Singapore with the aim to characterize the underlying mutations, which would contribute towards clinical applications such as confirmation of clinical diagnosis, genetic counseling and identification of applicable molecular therapies for these patients.
2 MATERIALS AND METHODS
2.1 Patient cohort and mutation screening
This study included 145 anonymized unrelated male patients, who were clinically diagnosed as DMD (n = 125) or BMD (n = 20) by a team of experienced pediatric neurologists (PSL, SKHT) and pathologist (JC) based on family history and clinical features including serum CK levels, age of onset, age at loss of ambulation, calf muscle hypertrophy, Gower's sign, presence of cardiomyopathy, and electromyographic patterns, in addition to dystrophin immunostaining results from muscle biopsies, where available. Samples from all patients were collected with written informed consent and in accordance with the principles of the Declaration of Helsinki. The study was approved by the institutional review board of the National Healthcare Group of Singapore.
Some of these patients were previously studied using multiplex PCR for selected hotspot exons in DMD gene or analyzed by Sanger sequencing (Bennett et al., 2009; Lai et al., 1992; Lai et al., 2002; Loke, Poh, Lee, & Lai, 2009; Low et al., 1996; Tay et al., 2006). These previously reported cases were included in this study and reanalyzed since previous studies conducted using available technologies at that time could not detect duplications or the full extent of the type of mutations covering all 79 coding exons of the DMD gene. Identification of whole exon deletions and duplications was carried out with the MLPA DMD kit (SALSA MLPA KIT P034/P035 DMD/Becker, The Netherlands) following the manufacturer's protocol using genomic DNA isolated from peripheral blood of patients. All the identified deletions and duplications were analyzed for changes in the reading-frame using the online DMD exonic deletions/duplications reading-frame checker 1.9 (https://www.humgen.nl/scripts/DMD_frame.php). For samples with negative MLPA results, sequencing was carried out by Sanger method (n = 32) or by whole exome sequencing (n = 4). Sanger sequencing of all the exons and exon-intron boundaries in the DMD gene was performed following standard methods (Aartsma-Rus et al., 2016; Bennett, den Dunnen, O'Brien, Darras, & Kunkel, 2001). Whole exome sequencing (WES) was carried out on Illumina Hiseq 4000 platform (Illumina, San Diego, CA) at a mean read depth of 100x. Genomic DNA libraries were prepared by Agilent SureSelect Human All Exon V5 Kit (Agilent Technologies, Santa Clara, CA) in accordance with the manufacturer's protocol. Variants were called via Genome Analysis Toolkit (GATK3.4) pipeline and annotated via ANNOVAR. The alignment files were analyzed via UCSC browser (Kent et al., 2002) to determine the coverage of every exon of the DMD gene and the quality of the reads.
2.2 Variant analysis
All variants identified from Sanger and exome sequencing were analyzed using in-house bioinformatics pipeline. Briefly, variants were curated against known population databases, namely—exon sequencing projects (ESP) (http://evs.gs.washington.edu/EVS/), exome aggregation consortium (ExAC), 1,000 genomes project database, genome aggregation database (gnomAD), and Kaviar (Auton et al., 2015; Glusman, Caballero, Mauldin, Hood, & Roach, 2011; Karczewski et al., 2019; Lek et al., 2016). Other resources used for curation were mutation databases—Leiden open variation database (LOVD) (https://databases.lovd.nl/shared/genes/DMD), Emory database (http://functionalvariants.emory.edu/database/index.html), and clinical databases—Clinvar (https://www.ncbi.nlm.nih.gov/clinvar/) and Clinvitae (http://clinvitae.invitae.com/). All variants were assessed for deleteriousness via predictive algorithms—SIFT (Sim et al., 2012), Polyphen (Adzhubei et al., 2010), mutation taster (Schwarz, Cooper, Schuelke, & Seelow, 2014), combined annotation dependent depletion (CADD) (Kircher et al., 2014), genomic evolutionary rate profiling (GERP) (Cooper et al., 2005), and PhyloP (Pollard, Hubisz, Rosenbloom, & Siepel, 2010) for functional impact arising from the identified sequence variants. All identified small mutations were classified into five categories as pathogenic, likely pathogenic, variants of unknown significance (VUS), benign or likely benign following the ACMG guidelines (Richards et al., 2015).
3 RESULTS
3.1 Mutation spectrum
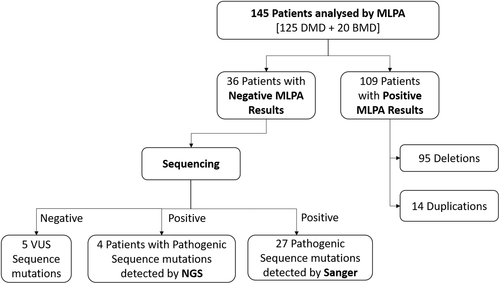
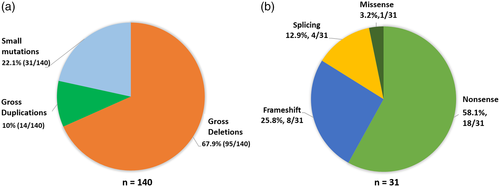
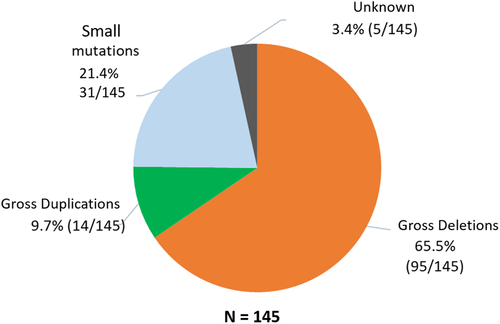
We applied a combinatorial approach of using MLPA followed by either Sanger sequencing or WES to detect candidate mutations in the DMD gene across 145 patients (Figure 1). Causal mutations in DMD gene were identified in 140 (96.6%) of the patients. Among the distribution of mutations in the 140 patients, the most frequent type of mutations were large deletions (67.9%; 95/140), followed by pathogenic or likely pathogenic small mutations (22.1%; 31/140), and duplications (10%; 14/140) (Figure 2a). We could not detect likely causative DMD variants in the remaining 3.4% (5/145) cases (Figure 3). Whole exome sequencing of DMD gene was performed in four recent cases (negative for deletion or duplication mutation from MLPA analysis) and causal variants were identified in all patients. All small mutations identified from both Sanger and exome sequencing were classified based on ACMG guidelines (Tables 1 and S1). Overall, 31 patients harbored either a pathogenic or a likely pathogenic small mutation in DMD gene as detected by either Sanger sequencing or WES. Among these mutations, 58.1% (18/31) were stop-gains, 25.8% (8/31) were frameshifts, 12.9% (4/31) were splicing and 3.2% (1/31) were missense mutations (Figure 2b). A total of nine novel DMD variants were identified in this study, six of which were likely pathogenic truncating mutations (one nonsense, four frameshifts, and one splicing). Three missense variants were classified as VUS and were not likely to be causative for the given disease phenotype (Table S2).
| Case ID | Phenotype | Mutation type | Exon/intron | Codon change | Protein change | ACMG classification | Reference |
|---|---|---|---|---|---|---|---|
| Case 137 | DMD | Missense | 2 | c.77A>G | p.Asn26Ser | Likely pathogenic—PS4, PM1, PM2, PP3, BP1 | (Wang et al., 2019; Yang et al., 2013) |
| Case 138 | DMD | Stopgain | 6 | c.433C>T | p.Arg145Ter | Pathogenic—PVS1, PS4, PM2 | (de Almeida et al., 2017; Flanigan et al., 2009; Guo et al., 2015; Nigro et al., 1994; Okubo et al., 2016; Yasuhiro Takeshima et al., 2010; Taylor et al., 2007) |
| Case 111a | DMD | Frameshift insertion | 7 | c.544dupG | p.Asp182GlyfsTer4 | Likely pathogenic—PVS1, PM2 | Novel |
| Case 112 | DMD | Frameshift substitution | 7 | c.616_617TGA | Likely pathogenic—PVS1, PM2 | Novel | |
| Case 113 | DMD | Stopgain | 7 | c.C568T | p.Gln190Ter | Pathogenic—PVS1, PS4, PM2 | (Flanigan et al., 2009; Magri et al., 2011) |
| Case 114 | DMD | Stopgain | 7 | c.583C>T | p.Arg195Ter | Pathogenic—PVS1, PS4, PM2 | (Buzin et al., 2005; Cho et al., 2017; Fajkusová et al., 2001; Flanigan et al., 2009; Guo et al., 2015; Lim et al., 2011; Mendell et al., 2001; Okubo et al., 2016; Prior & Bridgeman, 2005; Todorova et al., 2008) |
| Case 115 | DMD | Stopgain | 8 | c.829C>T | p.Gln277Ter | Pathogenic—PVS1, PM2, PS4 | (Bennett et al., 2009; de Almeida et al., 2017; L. Wang et al., 2019) |
| Case 117 | DMD | Stopgain | 16 | c.1817delT | p.Leu606Ter | Likely pathogenic—PVS1, PM2 | Novel |
| Case 139 | DMD | Stopgain | 16 | c.1904C>G | p.Ser635Ter | Pathogenic—PVS1, PS4, PM2 | (Cho et al., 2017) |
| Case 118 | DMD | Stopgain | 20 | c.2530C>T | p.Gln844Ter | Pathogenic—PVS1, PM2, PP5 | UMD-DMD France mutations database record ID: 2819 (unpublished) |
| Case 119 | DMD | Frameshift insertion | 20 | c.2614dupA | p.Ile872AsnfsTer3 | Pathogenic—PVS1, PS4, PM2 | (Bennett et al., 2009) |
| Case 120 | DMD | Stopgain | 24 | c.3259C>T | p.Gln1087Ter | Pathogenic—PVS1, PS4, PM2 | (Nigro et al., 1994; Torella et al., 2010) |
| Case 121 | BMD | Stopgain | 25 | c.3358G>T | p.Glu1120Ter | Pathogenic—PVS1, PM2, PP5 | Clinvar variation ID: 587510, genomic research center (Shahid Beheshti University of Medical Sciences) |
| Case 122 | DMD | Stopgain | 26 | c.3500C>G | p.Ser1167Ter | Pathogenic—PVS1, PS4, PM2 | (Flanigan et al., 2009; Okubo et al., 2017a) |
| Case 123 | DMD | Stopgain | 30 | c.4150G>T | p.Glu1384Ter | Pathogenic—PVS1, PS4, PM2 | (Flanigan et al., 2009) |
| Case 124 | DMD | Frameshift deletion | 34 | c.4681_4682del | p.Glu1561LysfsTer14 | Likely pathogenic—PVS1, PM2 | Novel |
| Case 125 | DMD | Stopgain | 34 | c.4838G>A | p.Trp1613Ter | Pathogenic—PVS1, PM2, PP5 | Clinvar variation ID: 523468 |
| Case 126 | DMD | Frameshift deletion | 34 | c.4786_4787del | p.Lys1596GlufsTer5 | Pathogenic—PVS1, PM2, PP5 | EGL genetics EmVclass db (unpublished) |
| Case 127 | DMD | Stopgain | 37 | c.5266C>T | p.Gln1756Ter | Pathogenic—PVS1, PS4, PM2 | (Prior & Bridgeman, 2005) |
| Case 128 | DMD | Stopgain | 40 | c.5646C>G | p.Tyr1882Ter | Pathogenic—PVS1, PS4, PM2 | (Juan-Mateu et al., 2015) |
| Case 129 | DMD | Frameshift insertion | 46 | c.6644_6645insC | p.Glu2215AspfsTer8 | Pathogenic—PVS1, PS4, PM1,PM2 | (Rininsland & Reiss, 1994) |
| Case 130a | DMD | Frameshift deletion | 47 | c.6804_6807del | p.Lys2268AsnfsTer2 | Pathogenic—PVS1, PS4, PM2 | (Flanigan et al., 2009), Clinvar variation ID: 290564 |
| Case 131 | DMD | Stopgain | 55 | c.8038C>T | p.Arg2680Ter | Pathogenic—PVS1, PS4, PM2 | (Bennett et al., 2009) |
| Case 132 | DMD | Frameshift deletion | 56 | c.8355delG | p.Lys2785AsnfsTer39 | Likely pathogenic—PVS1, PM2 | Novel |
| Case 133 | DMD | Stopgain | 57 | c.8416C>T | p.Gln2806Ter | Pathogenic—PVS1, PS4, PM2 | (Okubo, et al., 2017, Taylor, Maroulis, et al., 2007) |
| Case 134 | DMD | Stopgain | 60 | c.8944C>T | p.Arg2982Ter | Pathogenic—PVS1, PS4, PM2 | (Daoud et al., 2009; Deburgrave et al., 2007; Flanigan et al., 2009; Guo et al., 2015; Magri et al., 2011; Mendell et al., 2001; Okubo, et al., 2017a; Roberts et al., 1992; Spitali et al., 2009; Taylor, Maroulis, et al., 2007; Wang et al., 2019; Xu et al., 2018) |
| Case 135 | DMD | Stopgain | 64 | c.9337C>T | p.Arg3113Ter | Pathogenic—PVS1, PS4, PM2 | (Buzin et al., 2005; Deburgrave et al., 2007; Flanigan et al., 2009; Guo et al., 2015; Okubo, et al., 2017a; Taylor et al., 2007; Wang et al., 2017; Wang et al., 2019) |
| Case 141 | BMD | Splicing | Intron 29 | c.4071 + 1G>T | Pathogenic—PVS1, PS4, PM2 | (Flanigan et al., 2009; Rudnik-Schöneborn, Weis, Kress, Häusler, & Zerres, 2008) | |
| Case 143a | BMD | Splicing | Intron 42 | c.6118-1G>A | Pathogenic—PVS1, PS4, PM2 | (Magri et al., 2011) | |
| Case 144a | DMD | Splicing | Intron 52 | c.7661-2A>G | Pathogenic—PVS1, PS4, PM2 | (Guo et al., 2015) | |
| Case 145 | BMD | Splicing | Intron 69 | c.10086 + 2dupT | Likely pathogenic—PVS1, PM2 | Novel |
- Note: All Variants were classified as per ACMG guidelines 2015 (Richards et al., 2015).
- a These cases were analyzed through WES.
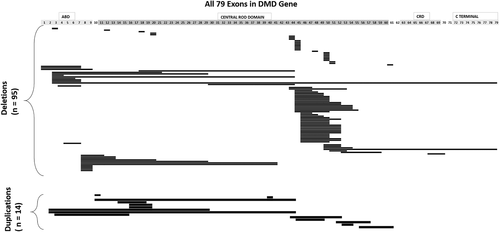
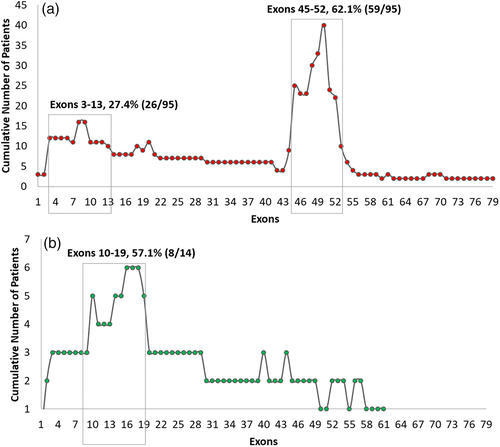
The distribution of exonic deletions and duplications among the patients show that 95 patients (65.5%) have gross deletions while large duplications were found in 14 (9.7%) patients (Figure 3). The deletions and duplications clustered around the two known proximal (exons 1 to 19) and distal (exons 44 to 55) hotspots (Figure 4). Two deletions extended into the region coding for the C-terminal protein with one of them starting from the proximal hotspot. Among the frequency of deletion per exon among patients, exons 3–13 in the proximal hotspot accounted for 27.4% (26/95) and exons 45–52 accounted for 62.1% (59/95) of the deletions (Figure 5). Exon 50 was found to be the most frequently deleted (n = 40) (Table S3). The most common region among the duplication mutations affected exons 10–19 in 57.1% (8/14) of cases. Novel duplications (exons 16–19 and exons 52–57) were identified in three patients (Table S4).
Reading frame analysis predicted 89 out-of-frame mutations and four in-frame mutations in 93 DMD patients while all 16 BMD patients were found to carry in-frame mutations (Tables S3 and S4). There was concordance to the reading frame rule in 96.3% (105/109) of these patients.
3.2 Predicted target exons for exon skipping
Single exon skipping is applicable to 77 patients with gross deletions and small mutations, while 26 patients will require multiple-exon skipping of more than one exon (Table 2). Single targeting of exon 51 is applicable to 18.9% of patients carrying deletions while targeting six exons individually (exons 8, 43, 45, 46, 51, and 53) would benefit 52.6% of these patients. Of the cases with point mutations, a total of 16 cases (51.6%) could be targeted by single exon skipping approach (Table 2). Multiple-exon skipping would be useful for 16.8% of deletions and 32.3% of patients with small mutations by targeting all nonsense and frameshift variants (Table 2). The exons targeted for skipping are shown in Tables S5 and S6. As remaining duplications span multiple exons, for which splicing strategies are often complex and unpredictable, candidates for exon skipping of these mutations are not reported (Aartsma-Rus et al., 2009; White et al., 2006).
| Strategy | Skipped exon(s) | Number of applicable patients with deletions | Frequency, n = 95 (%) | Number of applicable patients with sequence mutations | Frequency, n = 31 (%) |
|---|---|---|---|---|---|
| Single exon skipping | 7 | 1 | 1.1 | ||
| 8 | 4 | 4.2 | |||
| 11 | 1 | 1.1 | |||
| 16 | 2 | 6.4 | |||
| 17 | 1 | 1.1 | |||
| 19 | 2 | 2.1 | |||
| 22 | 1 | 1.1 | |||
| 24 | 1 | 3.2 | |||
| 25 | 1 | 3.2 | |||
| 26 | 1 | 3.2 | |||
| 30 | 1 | 3.2 | |||
| 34 | 3 | 9.7 | |||
| 37 | 1 | 3.2 | |||
| 40 | 1 | 3.2 | |||
| 43 | 2 | 2.1 | |||
| 45 | 7 | 7.4 | |||
| 46 | 7 | 7.4 | 1 | 3.2 | |
| 47 | 1 | 3.2 | |||
| 50 | 3 | 3.2 | |||
| 51 | 18 | 18.9 | |||
| 52 | 1 | 1.1 | |||
| 53 | 12 | 12.6 | |||
| 55 | 1 | 1.1 | 1 | 3.2 | |
| 56 | 2 | 6.4 | |||
| Total | 61 | 64.2 | 16 | 51.6 | |
| Multi-exon skipping | 6–7 | 10 | 10.5 | 1 | 3.2 |
| 6–8 | 4 | 12.9 | |||
| 7–8 | 1 | 1.1 | 1 | 3.2 | |
| 19–20 | 2 | 6.4 | |||
| 51–54 | 1 | 1.1 | |||
| 53–54 | 1 | 1.1 | |||
| 59–60 | 1 | 1.1 | |||
| 60–62 | 1 | 3.2 | |||
| 63–65 | 1 | 3.2 | |||
| 66–67 | 2 | 2.1 | |||
| Total | 16 | 16.8 | 10 | 32.3 |
4 DISCUSSION
We report an overall diagnostic rate of 96.6% (140/145) in DMD patients in Singapore using the methodologies described in this article. The deletion and duplication rates were 65.5 and 9.7% respectively. These frequencies are comparable to previous reports of 62–72.2% for deletions and 8.8–13.3% for duplications (Cho et al., 2017; Deepha et al., 2017; Guo et al., 2015; Vieitez et al., 2003; Rani et al., 2013; Juan-Mateu et al., 2015; Ma et al., 2018; Mohammed et al., 2018; Okubo, Goto, et al., 2017, ; Takeshima et al., 2010; Vengalil et al., 2017; Xu et al., 2018; Yang et al., 2013). The frequencies of large deletions and duplications may appear anomalous in some previous reports because of smaller cohort size or due to use of screening methods that are not able to detect the full spectrum of mutations (Rani et al., 2013; Vengalil et al., 2017). The combined mutation rate of 75.2% for gross deletions and duplications detected by MLPA in our population is similar to frequencies reported in other studies involving larger cohorts of Asian patients. These frequencies range from 70.56% among 1,053 patients (Yang et al., 2013) to 69.8% among 613 patients (Guo et al., 2015) in China and around 73% among 1,497 patients in Japan (Okubo et al., 2017a). Additionally, deletion of exon 50 was observed to be the most common exonic deletion in the Singaporean patient cohort, which was similar to other reported studies (Cho et al., 2017; Flanigan et al., 2009; Guo et al., 2015; Lai et al., 2002; Okubo et al., 2017a). Most of the gross deletion mutations in Singapore patients are commonly reported while three duplication mutations are novel.
Small mutations classified as pathogenic or likely pathogenic were observed in 21.4% (31/145) patients and in five cases, sequence variants were detected but did not fulfill the ACMG guidelines for pathogenicity. The presence of either pathogenic variants in non-coding regions of the DMD gene or complex genomic rearrangements affecting the DMD gene those were not detectable by the current techniques of WES and Sanger sequencing cannot be ruled out. It is also possible that other neuromuscular genes, which result in DMD-like phenotype could be involved and hence, other mutation screening methodologies like whole genome sequencing analysis, which can detect noncoding causal variants or novel genes, may be able to resolve such cases. This possibility cannot be ruled out as cases presenting overlapping phenotypes have previously been reported to be mistaken for DMD-like disease phenotypes (Dobrescu, Chelu, Tache, Purcaru, & Petrescu, 2015; Georgieva et al., 2005; Mudau, Essop, & Krause, 2016; Restagno et al., 1996). Hence, comprehensive mutation screening of DMD gene in local Singaporean patients with dystrophinopathies is crucial for providing precise genetic diagnosis.
In Singapore, routine molecular screening for DMD involves detecting the common deletions and duplications, which would be considered informative in 75.2% (109/145) of patients. In this study, a combination of MLPA and Sanger sequencing/WES detected 96.6% of patients with all types of DMD mutations. There is a growing possibility that the conventional diagnostic techniques in DMD such as MLPA and array comparative genome hybridisation (CGH) analysis for detecting copy number variants, and Sanger sequencing for detecting sequence variants will be replaced by next-generation sequencing technique given the decreasing costs of the latter (Aartsma-Rus et al., 2016). A limitation of the commonly used MLPA analysis is that it can only detect copy number mutations, deletions or duplications, and may yield a false positive result especially when single exon deletion is observed due to the presence of a polymorphism in the primer ligation site (Liu et al., 2018; Niba et al., 2014). Hence, for single exon deletions, a second confirmatory method of analysis is recommended. If next generation sequencing (NGS) approaches are used, the latter will not be needed as both sequence and copy number mutations are detected. For clinicians who are faced with a choice of different laboratory diagnostic approaches, the use of NGS panels is increasingly becoming more affordable and deals with the problem of DMD-like phenotypes. Although the turnaround time for MLPA and Sanger sequencing is less than the commercial clinical panel gene testing, which ranges between 2-6 weeks; a complete Sanger sequencing of DMD gene is tedious and requires around 94 PCR reactions followed by sequencing. Additionally, using a single NGS-based platform to simultaneously detect copy number variations (CNVs) and small mutations involving single nucleotide variations (SNVs) reduces the actual cost of diagnosis compared to doing a MLPA followed by a Sanger sequencing approach (Alame et al., 2016; Schofield et al., 2017; Wei et al., 2014). The future use of whole genome sequencing methodology can also identify structural gene rearrangements in addition to CNVs and SNVs.
However, there are still limitations to the wide scale application of NGS and a strong index of suspicion may sometimes require alternative methodologies such as a muscle biopsy for confirmation of diagnosis. Historically, muscle biopsies have been part of the initial work-up for diagnosis of neuromuscular disorders but there has been increasing use of genetic testing alone instead of using both genetic testing and muscle biopsy in diagnosis (Ryder et al., 2017). Nonetheless, muscle biopsies may still play an important role in cases where molecular genetic testing does not lead to definitive diagnosis (Suriyonplengsaeng et al., 2017).
The reading-frame rule which predicts the translational frame of a deletion or duplication and its correlation to either a DMD or a BMD phenotype is known to hold true in close to 90% or more of such cases (Aartsma-Rus, Van Deutekom, Fokkema, Van Ommen, & Den Dunnen, 2006; Flanigan et al., 2009; Guo et al., 2015; Juan-Mateu et al., 2015; Takeshima et al., 2010; Tuffery-Giraud et al., 2009; Vengalil et al., 2017; Yang et al., 2013). Exceptions to reading frame rule have been reported and attributed to several reasons such as site of the mutation or the presence of additional mechanisms, for example, ribosomal frame shift, cryptic promoter, alternate splicing, and RNA editing which maintains the reading frame (Aartsma-Rus et al., 2006; Gangopadhyay et al., 1992; Koenig et al., 1989). Other possible reasons for discordant genotype–phenotype correlations include tandem duplications or the presence of deletion breakpoints on exons itself (Guo et al., 2015; Wei et al., 2014). In this study, three deletion patients and one duplication patient in the Singapore cohort did not conform to the reading frame rule. The deletion cases spanned exons 3–44, 30–44, and 45–48, and were predicted to be in-frame but the patients clinically manifested as DMD. A possible explanation for this discrepancy of the exons 3–44 deletion is that in-frame deletions at the 5′ end of the gene which encodes for N-terminus of dystrophin including actin binding domain, as well as those including both N-terminal domain and the central rod domain have been known to be associated with a more severe DMD-like phenotype (Aartsma-Rus et al., 2016; Nicolas et al., 2012). In-frame deletion of exons 3–44 and exons 45–48 have been reported in four and two patients respectively who manifest DMD phenotype in the e-Dystrophin database of in-frame DMD mutations (http://edystrophin.genouest.org/). Deletion of exons 30–44 has also been previously been reported in a case of DMD (de Almeida et al., 2017) while duplication of exons 45–49 have been reported in three DMD patients (http://edystrophin.genouest.org/) and one patient with mild DMD phenotype (Pikó et al., 2009). Further studies at the mRNA transcript levels can clarify the underlying genetic mechanisms for the discrepancies for these cases.
A noteworthy observation in this study is the presence of a BMD patient with a nonsense mutation. This BMD patient carried a nonsense mutation in an in-frame exon 25. Previously, nonsense mutations in DMD gene have been associated with BMD phenotypes (Okubo et al., 2017b; Flanigan et al., 2011; Shiga et al., 1997). This has been attributed to skipping of the exon containing the nonsense mutation due to weak splice acceptor (3'ss) site and low exonic splicing enhancer density which leads to an in-frame deletion. Additionally, we observed a DMD patient with a missense mutation affecting the actin-binding domain (Exon 2, p.Asn27Ser). It has been previously reported that first 90 amino acids of the dystrophin tandem calponin homology domain is necessary for actin binding in vitro (Fabbrizio, Bonet-Kerrache, Leger, & Mornet, 1993). Ever since the first report of a DMD patient with missense mutation, it has been shown that missense mutations in actin binding domain one may cause loss of dystrophin function via protein instability and aggregation (Henderson, Lee, & Ervasti, 2010; Prior et al., 1993). It has also been observed that nearby mutations such as Lys18Asn and Leu54Arg are associated with very severe disease phenotype and each caused a small but significant fourfold decrease in actin-binding affinity (Henderson et al., 2010). Further biological studies are needed to clarify the basis underlying the unexpected phenotypes in our patients. Nonetheless, reports of such diverse mutation spectrum of DMD and BMD patients contribute toward further understanding on the molecular mechanisms of this disease.
Targeting of exons 8, 44, 45, 50, 51, 52, 53, and 55 individually has been predicted to help rescue reading-frame in roughly 4, 8, 13, 5, 15, 3, 9 or 2% of affected DMD patients, respectively (van Deutekom et al., 2001). We found 26.2% (38/145) of Singaporean dystrophinopathic patients carrying both deletions and point mutations to be eligible for therapeutic interventions targeting the skipping of exon 51 (Eteplirsen), 53 (Golodirsen/SRP-4053) and 45 (Casimersen/SRP-4045) using antisense nucleotides from Sarepta Therapeutics Inc. (USA). Further, multi-exon skipping may be beneficial to an additional 16.8% (n = 16) of local patients with deletion mutations, a frequency that is closely similar to observations from other populations (Aartsma-Rus et al., 2009; Béroud et al., 2007; Yokota, Pistilli, Duddy, & Nagaraju, 2007). It is also noteworthy that a significant proportion of our patients, that is, 12.4% (18/145) with nonsense mutations could potentially benefit from premature stop codon translational read-through therapies using aminoglycosides or small molecule compounds like Ataluren or Translarna™ (PTC Therapeutics, USA).
Notwithstanding the exciting developments using exon skipping and premature stop codon read-through therapeutics, it should be noted that both are transient forms of therapy and other approaches such as AAV mediated gene replacement or genome editing correction may offer more permanent correction of mutations in patients (Dickson, Roberts, Wells, & Fabb, 2002; Duan, 2016; Salmaninejad et al., 2018; Scott et al., 2002). Very promising results were recently reported in four patients showing robust restoration of gene expression from transduced microdystrophin (globenewswire.com/news-release/2018/10/03/1601085/0/en/Sarepta-Therapeutics-Announces-that-at-the-23rd-International-Congress-of-the-World-Muscle-Society-Jerry-Mendell-M-D-Presented-Positive-Updated-Results-from-the-Four-Children-Dosed.html). This strategy has the advantage of being non-mutation specific and requires just a single application of the drug to significantly ameliorate the histological and physiological signs of muscular dystrophy (Duan, 2018). Other permanent gene replacement therapies using CRISPR-based gene editing are currently being developed (Long et al., 2018). The latter, which is a mutation-specific approach, has been used to correct exon deletions, and duplications as well as sequence mutations (Lattanzi et al., 2017; Li et al., 2015; Young et al., 2016).
The latest standards of care for DMD updated in 2018 (Birnkrant et al., 2018a, 2018b, 2018c) recommends anticipatory diagnostic and therapeutic strategies for DMD patients especially with patients achieving longer survival ages. Hence, accurate and early genetic diagnosis of DMD/BMD is also important for disease management, which includes surveillance of possible cardiac, respiratory or orthopedic complications to ensure prompt interventions (Bushby et al., 2010).
Molecular diagnosis can also offer options such as carrier screening, prenatal diagnosis, preimplantation genetic diagnosis or enrolment into clinical therapeutic trials. Identifying the precise molecular defect in DMD is important for mutation-specific therapies such as exon skipping, premature stop codon read-throughs, and gene editing to be applied for patients. The ability to sub-classify patients into groups that can respond to specific targeted therapeutic approaches represents a paradigm for precision medicine in this disease. In conclusion, our findings provide the first insights into the dystrophinopathy mutation spectrum in Singapore and highlight the applicability of current therapeutic measures using exon skipping or nonsense read-throughs to local patients. This would further help in elucidating the molecular nature of the disease and guide long-term development plans for specific exon skipping and other drug therapies in the local context.
ACKNOWLEDGMENTS
We thank Ms Arthi Shanmugasundaram for assistance with the variant curation and Dr Monkol Lek for constructive review of the manuscript. We would like to acknowledge the past and current laboratory members of HUMGEN Laboratory who had provided the technical support for the experiments and thank the funding agencies (National Medical Research Council and Biomedical Research Council of Singapore) for supporting the projects that generated the data for this study. We dedicate this article to all our DMD and BMD patients who remain anonymous in this study but whose personal journeys and odysseys have become part of our quest in looking for improved diagnostics and therapeutics for this disorder.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
Biographies

Swati Tomar is a Research Fellow at the Department of Paediatrics at National University of Singapore. She is a molecular geneticist with expertise in next generation sequencing for inherited neuromuscular disorders and cancers. Her work focuses on unravelling causative mutations in early-onset rare genetic disorders in children.

Vikaesh Moorthy is a third-year medical student at the Yong Loo Lin School of Medicine, National University of Singapore. He is currently actively engaged in both clinical and laboratory research with a keen interest in congenital neuromuscular disorders and orthopedic pathologies.

Raman Sethi is a Research Associate working at Department of Paediatrics, Yong Loo Lin School of Medicine, National University of Singapore. He is a bioinformatician with research interest in Genomics and Computational Biology.

Josiah Chai is Head and Senior Consultant Neurologist at the Department of Neurology, National Neuroscience Institute in Singapore. He has clinical and research interests in neuromuscular diseases, electromyography and muscle histopathology.

Low Poh Sim is Professor of Pediatrics, Yong Loo Lin School of Medicine, National University Hospital in Singapore. Her research interests are in seizure disorder, neuromuscular diseases and developmental disorders.

Stacey Tay is an Associate Professor at the Department of Paediatrics, Khoo Teck Puat-National University Children's Medical Institute, National University Health Systems, Singapore. She oversees the running of multidisciplinary care clinics for neuromuscular patients. Her research interests are in neuromuscular disorders and neurogenetics.

Poh-San Lai is Associate Professor at the Department of Paediatrics, Yong Loo Lin School of Medicine, National University of Singapore and Khoo Teck Puat-National University Children's Medical Institute (KTP-NUCMI), National University Health Systems, Singapore. She heads the Human and Molecular Genetics Lab that first developed genetic tests for Duchenne muscular dystrophy in the country. Her research interest extends to both rare diseases and common conditions. She is Principal Investigator of several projects related to neuromuscular disorders and congenital diseases.




