Refining critical regions in 15q24 microdeletion syndrome pertaining to autism
Funding information: National Natural Science Foundation of China, Grant/Award Number: 81671362; Science and Technology Project of Jinan, Grant/Award Number: 201602194
Abstract
Chromosome 15q24 microdeletion syndrome is characterized by developmental delay, facial dysmorphism, hearing loss, hypotonia, recurrent infection, and other congenital malformations including microcephaly, scoliosis, joint laxity, digital anomalies, as well as sometimes having autism spectrum disorder (ASD) and attention deficit hyperactivity disorder. Here, we report a boy with a 2.58-Mb de novo deletion at chromosome 15q24. He is diagnosed with ASD and having multiple phenotypes similar to those reported in cases having 15q24 microdeletion syndrome. To delineate the critical genes and region that might be responsible for these phenotypes, we reviewed all previously published cases. We observe a potential minimum critical region of 650 kb (LCR15q24A-B) affecting NEO1 among other genes that might pertinent to individuals with ASD carrying this deletion. In contrast, a previously defined minimum critical region downstream of the 650-kb interval (LCR15q24B-D) is more likely associated with the developmental delay, facial dysmorphism, recurrent infection, and other congenital malformations. As a result, the ASD phenotype in this individual is potentially attributed by genes particularly NEO1 within the newly proposed critical region.
1 INTRODUCTION
Chromosome 15q24 microdeletion syndrome is a rare syndrome (Sharp et al., 2007), characterized by distinct phenotypes of global developmental delay, intellectual disability, facial dysmorphisms, congenital malformations of the hands and feet, eye, ear and genitalia, and joint laxity. The phenotypes are highly variable across different deletion carriers with >75% patients presenting facial features, developmental delay/intellectual disability, significant speech delay and hypotonia, 50–75% patients manifesting eye abnormalities, most frequently strabismus, digital anomalies, ear anomalies; 25–50% showing joint laxity, hearing loss, abnormalities on brain magnetic resonance imaging (MRI), hypospadias, and/or micropenis; 10–25% having seizures, sensorineural hearing loss, and conductive hearing loss; <10% featuring heart defects, diaphragmatic hernia, bowel atresia, and Pierre Robin syndrome (Mefford et al., 2012a, 2012b). It was first defined in 2007, with many more cases now reported and the prevalence assessed to be 3:10,000–4:10,000 ([Klopocki et al., 2008; Marshall et al., 2008; Kiholm Lund, et al., 2008; Andrieux et al., 2009; El-hattab et al., 2009; Masurel-Paulet et al., 2009, Van Esch et al., 2009; El-Hattab et al., 2010; McInnes et al., 2010; Ng et al., 2011; Brun et al., 2012; Mefford et al., 2012a, 2012b; Narumi et al., 2012; Gao et al., 2014; Samuelsson et al., 2015; Siu etal.,2016; Palazon-Bru et al., 2016; Romano et al., 2017; Ahram et al., 2017; Huynh et al.,2017; Dai et al., 2017; Wang et al., 2019]). Most of 15q24 deletions were found between 1.7 and 6.1 Mb in size with breakpoints localized to the clusters of five low copy regions (LCRs) from centromere to telomere, which were identified and designated as LCR15q24A (BP4), B (BP1), C, D (BP2), and E (BP3). Out of the cases, a few manifesting an additional phenotype of autism spectrum disorder (ASD; Ahram et al., 2017; McInnes et al., 2010; Mefford et al., 2012a, 2012b; Siu et al., 2016) carried mostly atypical deletions at 15q24 lying outside the critical region (Huynh et al., 2018; McInnes et al., 2010).
ASD is a vastly heterogeneous group of highly heritable neurodevelopmental disorders, characterized by impairments in reciprocal social interaction and communication, restricted and repetitive behaviors and interests, affecting ~1% of the population with the age of onset usually before 3 years (Carter & Scherer, 2013; Hoang, Cytrynbaum, & Scherer, 2018; Zhang et al., 2018). Various genetic mutations have been known to be implicated in ASD, ranging from chromosome abnormalities and copy-number variations (CNVs) to single-nucleotide variations (SNVs; Liu et al., 2015; Mak et al., 2017; Yuen et al., 2017). In 2007, the Autism Genome Project Consortium proposed the location of 15q23-25.3 would be the autism susceptibility locus by linkage and CNV analyses of 1,181 families with at least two ASD patients (Autism Genome Project Consortium, et al., 2007). Since then, more autistic patients have been reported carrying the 15q24 deletions (Ahram et al., 2017; Marshall et al., 2008; McInnes et al., 2010; Mefford et al., 2012a, 2012b; Siu et al., 2016). Here, we reported a de novo 2.58 Mb deletion at 15q24.1-q24.2 in a boy with ASD, and other phenotypes overlapped with those previously reported cases of 15q24 microdeletion syndrome. This is one case of a family-based study with the purpose of identifying genetic variants related to ASD. We reviewed all the available cases with 15q24 CNVs, focusing on microdeletions located at 15q24 in the published literature, DECIPHER (Firth et al., 2009) and ClinGen (Rehm et al., 2015; Solomon, Nguyen, Bear, & Wolfsberg, 2013) databases. We observed a second critical region of 650-kb interval between LCR15q24A and 15q24B that might contribute genetically to the ASD often found in this syndrome.
2 MATERIALS AND METHODS
The Review Board of Qilu Children's Hospital of Shandong University has approved this study. The patient's parents consented to this study.
2.1 Clinical samples and genomic DNA extraction
Peripheral blood samples from the patient and his healthy parents were obtained. Genomic DNA was extracted using DNAeasy Blood and Tissue Kit (QIAGEN, GmBH, Germany) following the manufacturer's instructions. Potential RNA contamination was removed by RNaseA (QIAGEN, GmBH, Germany). The DNA was quantified using NanoDrop ND-1000 spectrophotometer (Thermo Fisher, Waltham, MA).
2.2 Genome-wide SNP array and data analysis
The samples were genotyped using the Affymetrix Human Genome-Wide SNP Array 6.0. DNA digestion, ligation, fragmentation, labeling, hybridization, staining, and scanning were performed following the manufacturer's protocol (Affymetrix, Santa Clara). The data were analyzed with Command Console 3.1(Affymetrix, Santa Clara). The QC call rates of the samples are greater than 96.33%. The Database of Genomic Variants (MacDonald et al., 2014) was used as a reference of the structural variations identified in healthy human samples. Real-time quantitative PCR with the SYBR Premix Ex Taq II PCR reagent kit (TakaRa Bio, Dalian, China) was utilized with two sets of primers (F1: TACTGAAACCACCTGCATTCC and R1: TGTATCCTGCTCAACAATACCG; F2: AAACCAGCTCAGTGGAAATTG and R2: CGAGGAATTAAGAGCTGCAAG) for validation of the CNV. The genomic coordinates used are based on Human Genome Build GRCh37/hg19.
2.3 Identification of the minimal critical region
To define the critical region on chromosome 15q24, we curated all previously published cases in literature and from public clinical CNV databases such as DECIPHER (https://decipher.sanger.ac.uk/) and ClinGen (https://www.clinicalgenome.org/). The minimal critical locus defined as the region involving the maximum number of CNVs. The OMIM genes with only coding regions implicated in the critical region were analyzed for the potential roles. All the patients with available phenotypes from the literatures were summarized and the prevalence of the different phenotypes was counted. The clinical characteristics were then compared for the cases with overlapping deletions as similar size and loci as this case of ASD.
3 RESULTS
3.1 A de novo 15q24 microdeletion identified in the case diagnosed with ASD
A 2.58 Mb heterozygous deletion (chr15:72,964,144–75,544,822) was identified in a boy with ASD, mild developmental delay, severe speech problem, and hyperactivity. No additional rare CNVs found. The deletion located between 15q24A and 15q24C, disrupting 49 genes in total, of which nine (BBS, NEO1, HCN4, EDC3, MP1, SEMA7A, CYP11A1, CPLX3, STRA6, and ADPGK) were predicted to be possibly associated with ASD, neurodevelopment and congenital anomalies. Parental analysis revealed that the deletion was de novo in the patient. The real-time quantitative PCR confirmed the result.
3.2 Clinical description
The patient was a 2.5 year-old boy, the first child of healthy unrelated Chinese ethnicity parents, and was born by Caesarean section at term with weight 3.45 kg. His father and mother looked healthy and both of them denied any family history of neuropsychiatric disease. They were 28 and 26 years old at the time of his birth. He sat without support at 8 months, and was still unable to crawl. He started to walk at 18 months and distinguish family from strangers at nearly same time. He was referred to the Pediatric Health Care Institute for significant problems of speech and behaviors at 2.5 years. He was nonverbal vocabulary with only grunting sounds involuntarily and expressed his requirement by pointing and gestures. He always avoided eye contact with people without facial expression and initiative response, while manifested uncontrollable hyperactivity with uncoordinated movement and stereotypical behaviors like repetitive pacing, rocking in circles aimlessly. He had no obviously facial abnormality, but had pes planus and pigeon-toed in his left foot and a hearing loss in his left ear. His EEG, brain MRI, and neurological exams were normal. His development was tested via Gesell Developmental Observation-Revised, demonstrating his language/cognition, adaptation, motor skills, and personal-social interaction were significantly delayed. His ASD was diagnosed according to DSM-IV (Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, Chinese translation) criteria. At last investigation, he was 6-year-old and could only speak two words like mama and papa though he had taken a special language training program. The clinical features of this patient were compared to those 44 previously reported cases with available clinical phenotypes (Table 1). Further comparison analysis of genotypes and phenotypes was performed for this case and five previous reported patients with similar sizes and positions of deletions (Table 2).
| Phenotype | First region, no. (%) | Second region, no. (%) | Both regions, no. (%) | Out of both region, no. (%) |
|---|---|---|---|---|
| Male/female | 11/4 (33.3) | 12/4 (35.6) | 8/3 (24.4) | 2/1 (6.7) |
| Age at diagnosis <3 years | 5 (11.1) | 3 (6.7) | 3 (6.7) | 0 (0) |
| Inheritance of CNVs | 15 (33.3) | 16 (35.6) | 11 (24.4) | 3 (6.7) |
| De novo | 10 (22.2) | 14 (31.1) | 9 (20.0) | 2 (4.4) |
| Inheritance | 2 (4.4) | 1 (2.2) | 1 (2.2) | 0 (0) |
| Unknown | 3 (6.6) | 1 (2.2) | 1 (2.2) | 1 (2.2) |
| Developmental delay | 15 (33.3) | 16 (35.6) | 11 (24.4) | 3 (6.7) |
| Speech delay | 0 (0) | 8 (17.8) | 3 (6.7) | 0 (0) |
| Growth | 24 (53.3) | 15 (33.3) | 7 (15.6) | 5 (11.1) |
| Low birth weight | 9 (20.0) | 6 (13.4) | 1 (2.2) | 2 (4.4) |
| Short stature | 8 (17.8) | 3 (6.6) | 2 (4.4) | 2 (4.4) |
| Feeding difficulty | 5 (11.1) | 2 (4.4) | 2 (4.4) | 0 (0) |
| Obesity | 2 (4.4) | 4 (8.9) | 2 (4.4) | 1 (2.2) |
| Facial dysmorphism | ||||
| Face | 12 (26.7) | 8 (17.8) | 10 (22.2) | 1 (2.2) |
| High anterior hairline | 5 (11.1) | 4 (8.9) | 3 (6.7) | 1 (2.2) |
| Long face | 3 (6.7) | 3 (6.7) | 4 (8.9) | 0 (0) |
| Facial asymmetry | 4 (8.9) | 1 (2.2) | 3 (6.7) | 0 (0) |
| Eye | 29 (64.4) | 19 (42.2) | 18 (40.0) | 4 (8.9) |
| Downslanting palpebral fissures | 6 (13.3) | 3 (6.7) | 5 (11.1) | 1 (2.2) |
| Sparse eyebrows | 9 (20.0) | 2 (4.4) | 3 (6.7) | 1 (2.2) |
| Epicanthus | 5 (11.1) | 4 (8.9) | 5 (11.1) | 2 (4.4) |
| Strabismus | 4 (8.9) | 4 (8.9) | 2 (4.4) | 0 (0) |
| Hypertelorism | 4 (8.9) | 4 (8.9) | 2 (4.4) | 0 (0) |
| Nystagmus | 1 (2.2) | 2 (4.4) | 1 (2.2) | 0 (0) |
| Nose | 9 (20.0) | 4 (8.9) | 4 (8.9) | 0 (0) |
| Broad nasal base | 4 (8.9) | 2 (4.4) | 1 (2.2) | 0 (0) |
| Depressed nasal bridge | 3 (6.7) | 2 (4.4) | 1 (2.2) | 0 (0) |
| High nasal bridge | 2 (4.4) | 0 (0) | 2 (4.4) | 0 (0) |
| Ear | 11 (24.4) | 13 (28.9) | 9 (20.0) | 1 (2.2) |
| Ear abnormalities | 8 (17.8) | 9 (20/0) | 6 (13.3) | 0 (0) |
| Hearing loss | 3 (6.7) | 4 (8.9) | 3 (6.7) | 1 (2.2) |
| Mouth | 21 (46.7) | 15 (33.3) | 10 (22.2) | 2 (4.4) |
| Long smooth philtrum | 8 (17.8) | 3 (6.7) | 5 (11.1) | 1 (2.2) |
| Palate abnormalities | 5 (11.1) | 4 (8.9) | 1 (2.2) | 0 (0) |
| Full lower lip | 5 (11.1) | 2 (4.4) | 2 (4.4) | 0 (0) |
| Small mouth | 2 (4.4) | 4 (8.9) | 2 (4,4) | 1 (2.2) |
| Dental problems | 1 (2.2) | 2 (4.4) | 0 (0) | 0 (0) |
| Genital system | 4 (8.9) | 6 (13.3) | 6 (13.3) | 2 (4.4) |
| Hypospadias | 3 (6.7) | 2 (2.2) | 3 (6.7) | 1 (2.2) |
| Microphallus | 1 (2.2) | 2 (2.2) | 2 (4.4) | 0 (0) |
| Cryptorchidism | 0 (0) | 2 (2.2) | 1 (2.2) | 1 (2.2) |
| Digital anormalies | 17 (37.8) | 10 (22.2) | 5 (11.1) | 6 (13.3) |
| Thumb abnormalities | 3 (6.7) | 3 (6.7) | 1 (2.2) | 1 (2.2) |
| Brachydactyly short digit | 6 (13.3) | 3 (6.7) | 1 (2.2) | 3 (6.7) |
| Clinodactyly | 3 (6.7) | 0 (0) | 1 (2.2) | 0 (0) |
| Toe abnormalities | 5 (11.1) | 4 (8.9) | 2 (4.4) | 2 (4.4) |
| Recurrent infections | 7 (15.6) | 2 (4.4) | 4 (8.9) | 0 (0) |
| Behavior problems | 3 (6.7) | 13 (28.9) | 5 (11.1) | 2 (4.4) |
| ASD | 0 (0) | 6 (13.3) | 1 (2.2) | 0 (0) |
| ADHD/hyperactivity | 0 (0) | 3 (6.7) | 2 (4.4) | 0 (0) |
| Epilepsy | 0 (0) | 3 (6/7) | 1 (2.2) | 0 (0) |
| Happy personality | 3 (6.7) | 0 (0) | 0 (0) | 2 (4.4) |
| Intellectual disability | 0 (0) | 1 (2.2) | 1 (2.2) | 0 (0) |
| Skeletal system | 11 (24.4) | 7 (15.6) | 6 (13.3) | 1 (2.2) |
| Joint laxity | 6 (13.3) | 4 (8.9) | 4 (8.9) | 1 (2.2) |
| Scoliosis | 5 (11.1) | 3 (6.7) | 2 (4.4) | 0 (0) |
| Nervous system | 15 (33.3) | 15 (33.3) | 10 (22.2) | 3 (6.7) |
| Hypotonia | 8 (17.8) | 7 (15.6) | 5 (11.1) | 0 (0) |
| Microcephaly | 3 (6.7) | 1 (2.2) | 2 (4.4) | 2 (4.4) |
| Myelomeningocele | 1 (2.2) | 0 (0) | 0 (0) | 0 (0) |
| Abnormalities on brain MRI | 3 (6.7) | 7 (15.6) | 3 (6.7) | 1 (2.2) |
| Congenital heart disease | 4 (8.9) | 4 (8.9) | 2 (4.4) | 0 (0) |
| Other abnormalities | 3 (6.7) | 2 (4.4) | 4 (8.9) | 1 (2.2) |
| Hernias | 1 (2.2) | 1 (2.2) | 3 (6.7) | 1 (2.2) |
| Intestinal atresia | 2 (4.4) | 0 (0) | 0 (0) | 0 (0) |
| Imperforate anus | 0 (0) | 1 (2.2) | 1 (2.2) | 0 (0) |
| Case | 2012_Mefford_2 | 2012_Mefford_3 | 2012_Mefford_4 | 2012_Mefford_5 | 2012_Mefford_9 | 2017_Ahram | This case |
|---|---|---|---|---|---|---|---|
| Coordinate | 72,942,946–75,542,947 | 72,942,946–75,542,947 | 72,942,946–75,542,947 | 72,942,946–75,542,947 | 72,942,946–75,952,945 | 72,970,000–75,480,000 | 72,964,144–75,544,822 |
Size (Mb) Genes |
2.6 50 genes |
2.6 50 genes |
2.6 50 genes |
2.6 50 genes |
3.01 61 genes |
2.51 47 genes |
2.58 Mb 49 gene |
| Age at diagnosis | 6 years | 7 years | 12 years | 2.5 years | 6 years | 6 years | 2.5 years |
| Gender | Male | Male | Male | Male | Male | Female | Male |
| Motor development | Mild delay | Moderate delay | Mild delay, walked at 18 m | Moderate delay, walked at 30 m | Moderate delay, walked at 24 m | Mild delay, walked at 18 m | Mild delay, walked at 18 m |
| Speech problem | Significant speech delay | 2 words | Nonverbal | 20 words | Nonverbal | Nonverbal |
Nonverbal |
| Obesity | + | + | |||||
| Ear abnormality | + | + | + | ||||
| Hypotonia | + | + | + | + | |||
| Psychiatric | ADHD | ASD | ASD | Minimal eye contact | ASD | ASD, hyperactivity | ASD, hyperactivity |
| Foot/skeletal | Pet planus | Ligamentous laxity | Pet planus | Flat feet with significant pronation | Thenar hypoplasia | Pes planus and pigeon-toed |
- Abbreviations: ADHD, attention deficit hyperactivity disorder; ASD, autism spectrum disorder.
3.3 Identifying the critical region
To define the minimal critical region and genes that might contribute to the syndrome, we reviewed all the cases with isolated 15q24 deletions and duplications. In total, 46 deletions and 12 duplications were found including three embryos of miscarriage from literature, and an additional 16 deletions were from DECIPHER database, which were summarized in Tables S1–S3, separately. The clinical features were compared and the occurrence of each phenotype was calculated for all cases with 15q24 deletions except that case of embryo (Table 1). The CNV map of all cases was represented graphically in Figure 1 with two highlighted minimal critical regions. The first critical region is the one previously defined (chr15:74,500,000–75,700,000; 15q24B-D; first critical region) of a 1.2 Mb interval involving 52 (83.9%) cases of deletions and six (50.0%) cases of duplications. This region is gene-rich encompassing 39 genes including some OMIM genes (e.g., STRA6, CYP11A1, SEMA7A, ARID3B, CPLK3, CSK, and SIN3A; Magoulas & El-Hattab, 2012). This locus is more likely associated with other phenotypes of the syndrome excluding ASD. The second critical region is predicted from chr15: 73,010,000–73,660,000 (second critical region) covering 650-kb implicating 37 (59.7%) cases of deletions and three (25%) cases of duplications, including four genes (BBS4, ADPGK, NEO1, and HCN4), which presumably contributed to the phenotype of ASD of this syndrome.
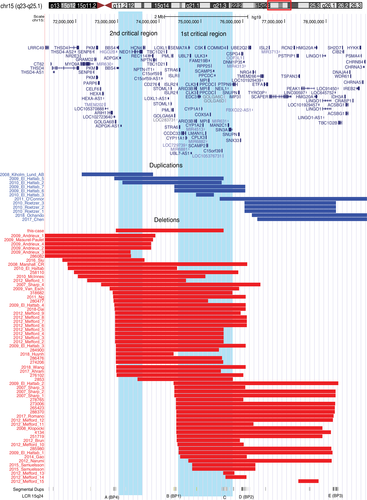
4 DISCUSSION
In this study, we proposed the second critical region of a 650-kb interval located between 15q24A and 15q24B based on cases study and review, which might contribute to the ASD phenotype of 15q24 microdeletion syndrome, while the first critical region of a 1.2-Mb span located between 15q24B and 15q24D defined previously is more likely associated with the other phenotypes of the syndrome, such as developmental delay, facial dysmorphism, recurrent infection, and congenital malformations (Magoulas & El-Hattab, 2012). Our case carried a de novo deletion of 2.58-Mb located between 15q24A and 15q24C presented the common phenotypes, such as severe speech problem, congenital anomalies, and additional ASD which is mostly attributed by the second critical region and the potential candidate gene of NEO1.
The smallest region of overlap has been predicted to be an approximately 1.2-Mb region between 15q24B and 15q24D as a majority of 19 patients with 15q24 deletions involved in this region (Magoulas & El-Hattab, 2012; McInnes et al., 2010; Mefford et al., 2012a, 2012b). So far, more cases with 15q24 deletions and duplications have been reported to present common clinical features (Roggenbuck et al., 2004; Miller et al., 2005; Schluth et al., 2005; Andrieux et al., 2009; El-Hattab et al., 2010; Roetzer et al., 2010; Cukier et al., 2011; Kim et al., 2011; O'Connor et al., 2011; Huynh et al., 2018; Ochando et al., 2018; Mefford et al., 2012b; Siu et al., 2016). To increase the number of cases, we searched all the literature, DECIPHER, and ClinGen databases for 15q24 deletions and duplications. By comparing the locus of CNVs linked to the clinical features, an interesting finding was noticed that the 25 cases carrying deletions located between 15q24B and 15q24E only covering the first critical region and excluding 15q24A and 15q24B did not present ASD phenotypes. Furthermore, all the ASD cases including our new patient in this study involved the deletions between 15q24A and 15q24B. Thus, we defined the minimal region within it as the second critical region, which is 650-kb in size including four genes, that is, BBS4, ADPGK, NEO1, and HCN4.
Previous studies have shown that homozygous loss-of-function mutations in some genes within first critical region of chr15q24 (see Figure 1) are associated with different congenital defects, such as STRA6 associated with Matthew-Wood syndrome presenting pulmonary agenesis/hypoplasia, congenital cardiac defect, microphthalmia/anophthalmia, diaphragmatic hernias, and urogenital malformations (Sadowski et al., 2017); ARID3B has been reported as a candidate gene associated with congenital cardiac malformations (Uribe et al., 2014); CYP11A, a cholesterol side chain cleavage enzyme associated with congenital lipoid adrenal hyperplasia (Horikawa, 2004); MPI related to congenital disorder of glycation and hyperinsulinemic hypoglycemia (Deeb & Al Amoodi, 2018); EDC3 related to autosomal recessive intellectual disability (Ahmed et al., 2015), indicating the haploinsufficiency of these genes might contribute to the corresponding phenotypes of the syndrome.
It has been known that some genes in first critical region of chr15q24 associated with the development of nervous system, such as SIN3A, SEMA7A, and CPLX3 (Cioli, Abdi, Beaton, Burnod, & Mesmoudi, 2014; Pasterkamp, Peschon, Spiggs, & Kolodkin, 2003; Witteveen et al., 2016), which suggests there might be potentially synergistic effects in these genes underlie the psychiatric phenotypes. In addition, two genes of NEIL1 and CSK associated with immunodeficiency, and T cell antigen receptor signaling (Lee et al., 2016; Manz et al., 2015; Romano et al., 2017; Sun et al., 2018; Zhang, Ren, Yang, & Ding, 2016).
By analyzing the clinical phenotypes of the cases implicated in 15q24, we defined the second critical region covering 650 kb located between 15q24A and 15q24B, encompassing four genes of BBS4, NEO1, HCN4, and ADPGK, which have been described to be associated with the atypical 15q24 deletion syndrome (Huynh et al., 2018; McInnes et al., 2010). BBS4 encoding Bardet-Biedl syndrome (BBS) 4 protein, is the causative gene of BBS that is an autosomal recessive ciliopathy characterized by obesity, polydactyly, renal malformation, rod-cone dystrophy, genital abnormalities, and cognitive disability (Ece Solmaz et al., 2015). Silencing of BBS4 promotes cell division and aberrant differentiation which results in adipose tissue formation underlying the BBS obesity (Aksanov, Green, & Birk, 2014).
NEO1, a membrane receptor, regulates pivotal developmental processes in the central nervous system of embryos involving interneuron migration, axon guidance, and cell death, which have been proved to contribute the autism pathophysiology (Fitzgerald, Cole, Hammond, Seaman, & Cooper, 2006; Matsunaga et al., 2004; Mawdsley et al., 2004; Wilson & Key, 2006). Siu et al. identified genetic changes in NEO1 gene from 2 out of 66 ASD patients and found the NEO1 played a critical role in cortical interneuron development by function experiment (Siu et al., 2016). Therefore, we proposed it might be a novel ASD-associated gene. HCN4 is a hyperpolarization-activated cyclic nucleotide gated channel 4 expressed predominantly in heart ventricle, atrium, and neurons and has been reported being associated with Sick Sinus Syndrome and cardiac abnormalities (Ishikawa et al., 2017; Milano et al., 2014). Recent studies showed it is highly expressed in habenula and thalamus, but is low expression level in amygdala, cortex, and hippocampus, which are known ASD associated regions. (Oyrer et al., 2019; Seo et al., 2015) and a loss of function mutation in HCN4 is associated with development of infantile epilepsy (Campostrini et al., 2018). ADPGK is ADP-dependent glucokinase and an alternative, glycolytic enzyme mediating generation of the oxidative signal whose downregulation inhibits oxidative signal and induces NF-kB-dependent gene expression, which makes T cell activation (Kaminski et al., 2012). We summarized the evidence of genes in the second region as Table 3. After studied, the phenotypes of the patients involved in this region, we predicted that the second critical region might be mainly responsible for behavior problems like ASD, but more studies are needed for the exact function of the potential critical genes within this region.
| Gene | pLI score | Gene function/detective phenotypes | mRNA expression in embryos | References |
|---|---|---|---|---|
| BBS4 | 0.00 | Bardet-Biedl syndrome; phenotypes of cardiovascular system, respiratory system, pigmentation and vision, digestive system, metabolism, integument, olfaction, craniofacial, mortality/aging, endocrine/exocrine gland, behavior/neurology, nervous system, reproductive system, liver/biliary system, adipose tissue, and so on. | Mainly expressed in testis (reproductive system), intestine (gastrointestinal tract), pancreas (endocrine system), ovary (reproductive system), blood (hematopoietic system). |
Iannaccone et al. (2005); Ece Solmaz et al. (2015); Aksanov et al. (2014); Li, Zhang, Jia, and Peng (2014). |
| NEO1 | 1.00 | Regulating pivotal developmental processes in the central nervous system of embryos involving interneuron migration, axon guidance, and cell death. Playing a critical role in cortical interneuron development. Diseases associated with NEO1 including hemochromatosis type2, cancers, and ASD. |
Mainly expressed in brain (nervous system) including third ventricle, hypothalamus, thalamus, striatum; neural tube including telencephalon, diencephalon, metencephalic alar plate, metencephalic basal plate; head mesenchyme; lung and trachea (respiratory system). |
Siu et al. (2016); Fitzgerald et al. (2006); Matsunaga et al. (2004); Mawdsley et al. (2004); Wilson and Key (2006). |
| HCN4 | 0.23 | Hyperpolarization-activated ion channel, contributes to the native pacemaker currents in heart that regulate the rhythm of heart beat, diseases-related including sick sinus syndrome, cardiac abnormalities, nervous system phenotype, mortality/aging, muscle phenotype, and so on. | Higher expressed in heart (cardiovascular system) including atrioventricular node, sinoatrial node, and early cardiomyocytes; brain (nervous system) including hypothalamus, thalamus, and cerebellum; eye (sensory organs), but lower expressed in amygdala, cortex and hippocampus. |
Milano et al. (2014); Ishikawa et al. (2017); Seo et al. (2015); Oyrer et al. (2019); Campostrini et al. (2018). |
| ADPGK | 0.02 | Encoding a ADP-dependent glucokinase mediating generation of the oxidative signal and play a role in glycolysis possibly during ischemic conditions. | Mainly expressed in blood and immune system like lymph node; internal like stomach, spleen; and lung; lower expressed in nervous system like cerebral cortex, heart, and kidney. |
Kaminski et al. (2012); Ronimus and Morgan (2004). |
- Abbreviation: ASD, autism spectrum disorder.
This study presents additional evidence of 15q24 microdeletion syndrome with clinical and molecular findings in a patient with ASD. Thorough review of the cases and this new patient suggests that the second critical region of 15q24 and genes, particularly NEO1, play an etiologic role on the ASD phenotype of the syndrome.
ACKNOWLEDGMENTS
The project is financially supported by Science and Technology Project of Jinan (201602194) and National Natural Science Foundation of China (81671362). The authors are grateful to the patient and his family for their contribution to the project. The authors would like to thank The Centre for Applied Genomics at the Hospital for Sick Children and the University of Toronto McLaughlin Centre. S.W.S. holds the GlaxoSmithKline-Canadian Institutes of Health Endowed Chair in Genome Sciences at the Hospital for Sick Children and University of Toronto.
CONFLICT OF INTEREST
S.W.S. is on the Scientific Advisory Committee of Population Diagnostics and Deep Genomics, and Lineagen Corp. Licenses intellectual property relevant to autism testing held by the Hospital for Sick Children.
AUTHOR CONTRIBUTIONS
Y.L., S.W.S., and Z.G. conceived and designed the study. X.Y. and Y.L. conducted the experiments. Y.L., M.Z., and S.W.S. analyzed the data. Y.Z., R.D., D.Z., and Z.G. contributed clinical diagnosis of the patients. Y.L. and M.Z. wrote the manuscript.




