Exome sequences of multiplex, multigenerational families reveal schizophrenia risk loci with potential implications for neurocognitive performance
Abstract
Schizophrenia is a serious mental illness, involving disruptions in thought and behavior, with a worldwide prevalence of about one percent. Although highly heritable, much of the genetic liability of schizophrenia is yet to be explained. We searched for susceptibility loci in multiplex, multigenerational families affected by schizophrenia, targeting protein-altering variation with in silico predicted functional effects. Exome sequencing was performed on 136 samples from eight European-American families, including 23 individuals diagnosed with schizophrenia or schizoaffective disorder. In total, 11,878 non-synonymous variants from 6,396 genes were tested for their association with schizophrenia spectrum disorders. Pathway enrichment analyses were conducted on gene-based test results, protein-protein interaction (PPI) networks, and epistatic effects. Using a significance threshold of FDR < 0.1, association was detected for rs10941112 (p = 2.1 × 10−5; q-value = 0.073) in AMACR, a gene involved in fatty acid metabolism and previously implicated in schizophrenia, with significant cis effects on gene expression (p = 5.5 × 10−4), including brain tissue data from the Genotype-Tissue Expression project (minimum p = 6.0 × 10−5). A second SNP, rs10378 located in TMEM176A, also shows risk effects in the exome data (p = 2.8 × 10−5; q-value = 0.073). PPIs among our top gene-based association results (p < 0.05; n = 359 genes) reveal significant enrichment of genes involved in NCAM-mediated neurite outgrowth (p = 3.0 × 10−5), while exome-wide SNP-SNP interaction effects for rs10941112 and rs10378 indicate a potential role for kinase-mediated signaling involved in memory and learning. In conclusion, these association results implicate AMACR and TMEM176A in schizophrenia risk, whose effects may be modulated by genes involved in synaptic plasticity and neurocognitive performance.
1 INTRODUCTION
Schizophrenia is a debilitating mental disorder characterized by psychosis, cognitive impairment and neurophysiological features, often involving social and occupational dysfunction, with increased physical health problems and shorter lifespans (Mueser & McGurk, 2004). The onset of symptoms generally occurs in early adulthood, affecting approximately 1% of individuals globally (van Os & Kapur, 2009), with substantial burdens on national healthcare systems and economies. The rate of sustained recovery is modest, as pharmacological treatments are of limited efficacy for functional outcome and cognitive performance (Miyamoto, Miyake, Jarskog, Fleischhacker, & Lieberman, 2012), underscoring our need for greater understanding of schizophrenia's pathophysiological mechanisms.
Although a substantial genetic underpinning has been incontrovertibly demonstrated in twin, family and adoption studies, yielding heritability estimates of around 0.80 (Sullivan, Kendler, & Neale, 2003), much of the genetic liability of schizophrenia remains to be accounted for (Lee et al., 2012). Linkage screens, candidate gene studies, and genome-wide association studies (GWAS) have implicated various genetic loci in schizophrenia risk, like the major histocompatibility complex (MHC) region at locus 6p22, suggesting a highly polygenic architecture with potentially thousands of variants, both common and rare, mostly of small effect (Purcell et al., 2009).
To meet this challenge, some researchers have turned to next generation sequencing to comprehensively interrogate genetic variation from across the genome for disease association. Coupled with appropriate filtering protocols, this approach has led to the discovery of novel susceptibility genes for many traits, especially for Mendelian conditions (Ng et al., 2010). For more complex traits such as neuropsychiatric disorders, etiological insights have been gained, including the impact of de novo or rare mutational loads on schizophrenia risk (Girard et al., 2011), with disruptive variants showing enrichment among genes involved in calcium ion channels and post-synaptic complexes (Fromer et al., 2014).
Despite the mounting evidence of a polygenic burden, the identification of schizophrenia risk genes in population-based sequencing studies has been hindered by statistical power issues related to small effect sizes, even with the increasing enormity of the data sets. However, Timms et al. (2013) achieved a notable success in unraveling schizophrenia genetics by targeting multiplex families (i.e., with several schizophrenic individuals). Examining whole exome sequence variation, the authors reported on the co-segregation of rare, protein-altering variants in N-methyl-D-aspartate (NMDA) receptor genes, lending support for the glutamatergic hypofunction hypothesis of schizophrenia. A more limited look at exome sequence variants within a previously implicated linkage region in multiplex families implicated AMPA receptor trafficking, which also ties into glutamatergic function (Kos et al., 2016).
In this paper, we conduct exome sequencing in eight multiplex, multigenerational European-American families, including 23 individuals diagnosed with schizophrenia or schizoaffective disorder, in an effort to uncover new susceptibility genes and further our understanding of the biological basis of schizophrenia spectrum disorders. Through association testing and pathway enrichment analysis, we identify protein-altering variants in genes involved in fatty acid metabolism as potential risk factors, representing novel targets for schizophrenia treatment and research.
2 MATERIALS AND METHODS
2.1 Family samples
From eight pedigrees ascertained for the Multiplex-Multigenerational Genetic Investigation of Schizophrenia (MGI) study (Almasy et al., 2008), samples from 136 individuals were exome sequenced, ranging from 6 to 26 individuals per family, including 21 diagnosed with schizophrenia and two cases of schizoaffective disorder of the depressed type (SAD), based on Diagnostic and Statistical Manual of Mental Disorders, 4th Edition (DSM-IV) criteria. Ages range from 15 to 86 years (mean = 48.8, SD = 19.8), with 44.9% male. Among the affected individuals, comorbidity included substance abuse and dependence (alcohol, amphetamines; n = 7 cases), antisocial personality disorder, mild mental retardation, and mood disorder. The selection of samples to be sequenced was based on various criteria, foremost being affection status and its pedigree density, availability and quality of DNA samples, and extremity of neurocognitive measures, in particular, the Penn Conditional Exclusion Test (PCET) that assesses abstraction and mental flexibility, features that are mediated by synaptic plasticity and often compromised in schizophrenia (Kos et al., 2016). Among the eight families chosen for our study, a mean of four individuals per family (SD = 1.20) were diagnosed with schizophrenia or SAD (mean for the other MGI families is approximately 2.2, SD = 0.48), which includes the most densely affected family with six cases. Mean PCET efficiency scores for the 136 study samples is −0.50 (SD = 1.19), which is significantly more dysfunctional than the mean score for the MGI samples not selected (−0.21, SD = 1.03; p = 8.9 × 10−3), including other members belonging to the eight target pedigrees (0.10, SD = 0.98; p = 3.0 × 10−4). The European-American families from MGI were originally recruited through a proband diagnosed with schizophrenia, with at least one first-degree relative affected with schizophrenia or SAD. All first-, second-, and third-degree relatives of 15 years of age or older were invited to participate. The study was approved by the Institutional Review Board of each of the collaborating institutions (University of Pennsylvania, University of Pittsburgh, and Texas Biomedical Research Institute), with informed consent obtained from all participants after receiving a complete description of the study, and parental consent required for minors.
2.2 Clinical diagnosis
DSM-IV diagnoses were achieved through a consensus of the following: 1) Diagnostic Interview for Genetics Studies, v. 2.0; 2) Family Interview for Genetics Studies; and 3) reviews of available medical records. For each subject, lifetime best-estimate diagnoses were determined by two investigators that include both psychiatrists and psychologists, each blind to the familial relationships of the participants. Cases involving psychotic features, as well as any discrepancies between the two raters, were further reviewed, with inter-rater reliability at kappa >0.8. In total, 106 individuals from 43 multi-generational families were diagnosed with schizophrenia or SAD. At the time of assessment, 75% of cases were undergoing treatment, including anti-psychotics. In addition to schizophrenia, other psychiatric conditions were identified in the MGI families, including personality disorders, schizoaffective disorder bipolar type, and delusional disorder.
2.3 Penn conditional exclusion test (PCET)
To evaluate neurocognitive performance, MGI participants underwent a computerized test battery (Gur et al., 2001a,2001b). Abstraction and mental flexibility were assessed using the PCET (Kurtz, Ragland, Moberg, & Gur, 2004), requiring participants to select one of four shapes for exclusion based on a sorting principle. An efficiency score was calculated as the average z score for performance accuracy and speed.
2.4 Exome sequencing
Exome sequencing was performed on 136 samples using the Illumina TruSeq platform on the HiSeq 2000 (Illumina, San Diego, CA), yielding approximately 62 megabases (Mb) of sequence data per sample, with uniform coverage of 201,121 exons from 20,794 genes (for more details, see, Supplementary Methods). Using CASAVA 1.8 suite, FASTQ files of de-multiplexed paired sequencing reads of 100 bp were produced, which were mapped to the UCSC human genome reference assembly 19 (hg19) with BWA (v. 0.6.1) (Li & Durbin, 2009). To ensure consistency, the mapped reads were analyzed with SAMtools (v. 0.1.12a) (Li et al., 2009) and Picard (v. 1.56) [http://picard.sourceforge.net] to mark likely PCR duplicates. The sequence data were then genotype called and filtered for quality with the GATK (v. 1.6) software package (McKenna et al., 2010), with recalibrated confidence scores greater than or equal to Q30, and assigned variant quality scores (VQS) based on covarying sequence annotations (e.g., quality score normalized by allele depth, haplotype score, strand bias, and mapping quality). The output was 380,895 single nucleotide polymorphisms (SNPs) in VCF format, with functional annotation achieved with ANNOVAR (v. 2013Feb21) (Wang, Li, & Hakonarson, 2010). Non-synonymous variants with potential deleterious effects on protein function, as predicted by the algorithms SIFT (scores >0.95; categorized as “damaging”) and/or PolyPhen2 (scores >0.15; categorized as either “probably damaging” or “possibly damaging”), are the focus of the study. In total, 11,878 variants were selected, with VQS LOD ≥4.0, 100% call rate, and Hardy–Weinberg Equilibrium p-values > 0.001 (computed in SOLAR).
2.5 TaqMan genotyping of replication samples
Five missense variants were selected for follow-up genotyping: rs10941112 (AMACR) and rs10378 (TMEM176A), both of which show significant association with schizophrenia in our discovery data set, and three SNPs from the gene AMACR, rs2278008, rs2287939, and rs34677, two of which have been previously implicated in schizophrenia risk. In total, 456 individuals from 41 families were genotyped, with 49.3% male and ranging in age from 15 to 87 years (mean = 45.1, SD = 16.7), including 64 individuals diagnosed with schizophrenia or SAD (n = 6), with a mean of 1.41 cases (SD = 0.95) per family (range: 1–3). Genotyping was performed using the TaqMan allelic discrimination methodology on a Life Technologies QuantStudio 12 K Flex instrument. Assays included a combination of pre-designed TaqMan SNP Genotyping assays and custom TaqMan SNP Assays designed using the Custom TaqMan Assay Design Tool. Assays were prepared in a 384-well plate in a final volume of 5 µl and PCR performed on the QuantStudio as per the manufacturer's protocol. Genotype data was analyzed and results exported using the TaqMan Genotyper Software on the QuantsStudio 12 K Flex system.
2.6 GWAS data from the psychiatric genomics consortium
Significant SNP association results from the exome sequence data were compared to the most recent GWAS data generated for schizophrenia by the Psychiatric Genomics Consortium (PGC) (Ripke et al., 2014). The study involved meta-analysis of 49 case control samples and three family based association studies (up to 36,989 cases and 113,075 controls in total), predominantly of European ancestry, testing approximately 9.5 million SNPs after quality control and imputation. Diagnoses were determined with DSM-IV or ICD-10 (10th revision of the International Statistical Classification of Diseases and Related Health Problems) criteria.
2.7 mRNA profiling
Gene expression data were examined for AMACR and TMEM176A, the genes harboring the significant association signals observed at rs10941112 and rs10378, respectively. RNA transcription levels were assayed in 107 study samples from lymphoblastoid cell lines using the Illumina HT-12 Expression BeadChips, with preliminary data analysis performed using Illumina GenomeStudio Software. Raw signals were log transformed and standardized by z-scoring within individuals (cell lines) to reduce the influence of inter-individual differences stemming from variation in RNA quality. For each transcript, an inverse Gaussian transformation was performed on the residual expression scores prior to association testing.
Data from the Genotype-Tissue Expression (GTEx) project, a comprehensive atlas of correlations between genotypes and tissue-specific gene expression levels (The GTEx Consortium, 2013), were also examined for SNPs significantly associated with schizophrenia in the MGI samples. GTEx donors (n = 544) of any sex, race, and ethnic group are identified through low-post-mortem-interval (PMI) autopsy or organ and tissue transplant settings. Biospecimens for 53 tissue types are quantified for gene expression from massively parallel sequencing of RNA. Peripheral blood samples are collected and used as a source for whole-genome SNP and copy number variant (CNV) genotyping, and to establish lymphoblastoid cell lines.
2.8 Statistical analysis
Genetic association tests and heritability (h2) estimation were performed using the program SOLAR (Almasy & Blangero, 1998), based on maximum likelihood variance decomposition methods. Associations between protein-altering variants and diagnoses of schizophrenia and SAD, as well as PCET efficiency scores and gene expression data, were examined via measured genotype analyses (MGA), under a liability threshold model for discrete traits. This is a mixed effects variance components approach in which kinship relations are treated as random effects, parameterized by additive genetic variation (i.e., residual heritability), with SNPs defined as fixed effects. The likelihoods of models saturated for the effects of both kinship and SNP genotypes were compared to null models with SNP effects constrained to zero, representing a single degree of freedom test. p-values were adjusted using the tail-area based False Discovery Rate (FDR) approach, with q-values estimated using the fdrtool package in R. SNPs with q-values <0.1, a threshold considered as statistically significant, were also evaluated for cis effects on gene expression. Covariates included sex, age, age squared, and their interactions. Pairwise genetic interactions were investigated by expanding MGA models to include main effects of a secondary SNP genotype and its interaction with rs10941112 or rs10378.
Gene-based association tests were performed in the software KGG v. 4.0 using GATES, an extended Simes test that combines p-values of SNPs within a gene to obtain an overall p-value (Benjamini–Hochberg correction), which has shown robust power for detecting genes with one or a few independent causal variants (Li, Gui, Kwan, & Sham, 2011). Gene annotation was achieved in KGG using the RefGene database, with SNP LD based on 1000 Genomes haplotypes for European population samples. Pathway enrichment was assessed among top associated genes (GATES p < 0.01; n = 87 genes) by conducting Fisher's exact tests on these observed results, as well as permuted data representing 100,000 similar-sized gene lists randomized from 7,300 genes annotated to the 11,878 target SNPs, yielding empirical p-values, adjusted with the Bonferroni approach. Pathways defined by KEGG, Biocarta, Pathway Interaction Database (PID) and Reactome (n = 1,273) were used in the enrichment analyses. Protein-protein interactions (PPIs) were also investigated among the top associated genes (GATES p < 0.05; n = 359 genes) using the web resource STRING v. 10.0, based on curated experimental data from primary interaction sources, reporting on results with PPI confidence scores greater than 0.40 (Szklarczyk et al., 2015).
3 RESULTS
3.1 Exome sequence data
From the targeted 62 Mb, exome sequencing yielded an average of 35 reads per nucleotide variant, with an average of 88,922 SNPs called per individual (Table 1). In total, 380,895 SNPs were identified, with approximately half (50.6%) displaying derived (i.e., non-ancestral) allele frequencies <5.0%, and a novelty rate of 21.8% relative to dbSNP 137 (note: GATK quality metrics available in supplementary Figures S1 and S2). The transition: transversion (Ti:Tv) ratio is 2.03, consistent with ratios reported for genome-wide sequence data, although markedly less than the 3.0–3.5 range generally observed for exonic variation (DePristo et al., 2011). When broken down according to genic location and function, 26.9% of the sequence variants are exonic, of which 57.1% are rare and uncommon (frequencies <5.0%), and a Ti:Tv ratio of just 2.70, likely reflecting the presence of false positives and/or unknown sequencing context bias (DePristo et al., 2011). To better control for Type I errors, we applied stringent filters on the exonic variants, retaining 57,684 autosomal SNPs with VQS LOD ≥4.0 and no missing genotypes, as well as excluding any de novo mutations, producing a Ti:Tv ratio of 3.41. From this subset, 11,878 missense variants with potential deleterious effects were identified using the SIFT and PolyPhen2 scores (minimum MAF = 0.0074), which are the focus of this association study.
| Derived allele frequency ranges | ||||||
|---|---|---|---|---|---|---|
| Sequence data | n | Ti:Tv | Novelty (%) | <0.5% | 0.5–5.0% | >5.0% |
| All Sites | 380,895 | 2.03 | 21.8 | 12.7 | 37.9 | 49.4 |
| Exonic | 102,468 | 2.70 | 15.5 | 16.4 | 40.7 | 42.9 |
| Intronic | 135,534 | 1.92 | 22.4 | 10.7 | 36.8 | 52.5 |
| Splicing | 1,006 | 0.79 | 50.3 | 15.3 | 38.5 | 46.2 |
| UTR | 106,956 | 1.81 | 25.7 | 12.5 | 37.6 | 49.9 |
| Upstream | 6,145 | 1.42 | 27.9 | 9.5 | 34.2 | 56.3 |
| Downstream | 5,510 | 1.76 | 22.5 | 7.2 | 33.4 | 59.4 |
| ncRNA | 22,059 | 1.88 | 25.1 | 10.7 | 34.8 | 54.5 |
| Intergenic | 1,217 | 1.80 | 27.2 | 10.7 | 35.2 | 54.1 |
| Autosomal, exonic, VQS LOD ≥4* | 57,684 | 3.41 | 6.8 | – | 51.8 | 48.2 |
| Synonymous | 27,872 | 5.67 | 4.9 | – | 46.5 | 53.5 |
| Non-synonymous | 29,418 | 2.35 | 8.4 | – | 56.6 | 43.4 |
| Stop-gain | 336 | 1.87 | 21.4 | – | 72.0 | 28.0 |
| Stop-loss | 40 | 1.11 | 15.0 | – | 47.5 | 52.5 |
| Predicted deleterious effects** | 11,878 | 2.13 | 10.8 | – | 68.0 | 32.0 |
- Table summarizes the called SNPs for the exome sequence data. Listed are the transition: transversion (Ti:Tv) ratios, novelty rates according to dbSNP build 137, and frequency ranges of derived alleles (<0.5% represent singletons), based on UCSC human genome reference assembly 19 (hg19).
- * Variant Quality Score (VQS) LODs were computed with VariantRecalibrator in the GATK software, representing probabilities of called variants being true variants, based on a mixed Gaussian model of covarying sequence annotations trained with known SNP data (i.e., HapMap 3.3 and dbSNP 137). Variants with missing genotype data for any of the 136 study samples (n = 2,068), as well as de novo and fixed sites, are not included in this sequence data subset. Exonic functions of 18 variants were characterized by ANNOVAR as “unknown” are also not represented in the table statistics.
- ** Category represents those autosomal, exonic variants with VQS LODs greater than or equal to 4.0 with predicted effects on function based on the SIFT and PolyPhen2 algorithms, as annotated by ANNOVAR. Variants include only those with SIFT scores >0.95 (categorized as “damaging”) and/or PolyPhen2 scores >0.15 (categorized as either “probably damaging” or “possibly damaging”). These variants were the focus of the present study. Average number of reads per nucleotide is approximately 43, with an average of 1,752 variant sites observed per individual. More than two-thirds of these SNPs are uncommon, an enrichment that likely reflects the consequences of purifying selection on deleterious mutations (Tennessen et al., 2012).
3.2 Association results for schizophrenia risk
The top ten association results for schizophrenia (h2 = 0.84 ± 0.78; p = 8.6 × 10−3) are presented in Table 2. Genomic inflation is observed (λ = 1.28, Q–Q plot presented in supplementary Figure S3), however given the nature of the targeted sequence data (i.e., deleterious missense mutations with intragenic LD) and the high heritability and polygenic architecture of schizophrenia (Purcell et al., 2009), this is not unexpected (Yang et al., 2011). The top association (p = 2.10 × 10−5) is for a common SNP (MAF = 0.39), rs10941112 (nucleotide transition G524A; amino acid substitution G175D), located in exon 3 in the gene AMACR at locus 5p13 (Figure 1), with the minor G allele showing evidence of decreased susceptibility (β = −0.97 ± 0.45), which is significant after adjustment using the FDR approach (q-value = 0.073). All 23 of the affected individuals are carriers of the risk allele, A, with 18 homozygotes and 5 heterozygotes, producing an inflated allele frequency of 89.1% among cases. This starkly contrasts the 113 unaffected members, with a genotype ratio of 37:50:26 for AA: AG: GG, corresponding to a frequency of 54.9% for the A allele. Based on 1000 Genomes data, the risk allele ranges in frequency from 3.3% to 51.5% for African and European populations, respectively. Precomputed scores of the Combined Annotation-Dependent Deletion (CADD) algorithm, a “meta-annotation” tool for assessing the effects of mutations on protein function (Kircher et al., 2014), ranks rs10941112 in the top 0.063% of the most deleterious SNPs among the 8.6 billion possible substitutions in the hg19 human reference genome.
| Function prediction | Schizophrenia risk | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| dbSNP 137 | Chrom. | Position (bp) | Gene | MA | MAF | SIFT | PolyPhen2 | Beta (SE) | p-Value |
| rs10941112 | 5 | 34,004,707 | AMACR | G | 0.39 | Damaging | Possibly damaging | −0.97 (0.23) | 2.10 ×10−5* |
| rs10378 | 7 | 150,501,455 | TMEM176A | T | 0.18 | Damaging | Probably damaging | 1.14 (0.27) | 2.80 × 10−5* |
| rs35176475 | 4 | 108,575,955 | PAPSS1 | A | 0.015 | Tolerated | Possibly damaging | 4.64 (1.21) | 1.28 × 10−4 |
| – | 1 | 213,302,933 | RPS6KC1 | G | 0.0074 | Damaging | Probably damaging | 7.27 (2.07) | 4.51 × 10−4 |
| – | 12 | 2,968,388 | FOXM1 | A | 0.0074 | Damaging | Probably damaging | 7.13 (2.03) | 4.51 × 10−4 |
| rs34637584 | 12 | 40,734,202 | LRRK2 | A | 0.0074 | Damaging | Probably damaging | 7.23 (2.06) | 4.51 × 10−4 |
| rs201530919 | 12 | 57,922,150 | MBD6 | A | 0.0074 | Damaging | Probably damaging | 7.13 (2.03) | 4.51 × 10−4 |
| rs10455840 | 6 | 159,401,898 | RSPH3 | T | 0.0074 | Damaging | Possibly damaging | 7.24 (2.06) | 4.51 × 10−4 |
| rs36111323 | 11 | 101,359,750 | TRPC6 | A | 0.12 | Tolerated | Possibly damaging | −4.23 (1.24) | 6.26 × 10−4 |
| rs74053516 | 11 | 5,602,929 | OR52B6 | A | 0.051 | Tolerated | Possibly damaging | 1.55 (0.45) | 6.26 × 10−4 |
- Table lists the top ten association results for non-synonymous, potentially damaging exome variants with schizophrenia risk. SNP rs numbers are based on dbSNP build 137. Chromosome (Chrom.) and position correspond to hg19. Minor allele frequencies (MAFs) are based on maximum likelihood estimates that account for familial relationships. Predicted effects of amino acid changes on protein function are based on SIFT and PolyPhen2 scores assigned by ANNOVAR.17 Positive beta estimates (standard error in parentheses) for schizophrenia correspond to increased, additive risk for the minor allele (MA).
- * False Discovery Rate (FDR) q = 0.073.
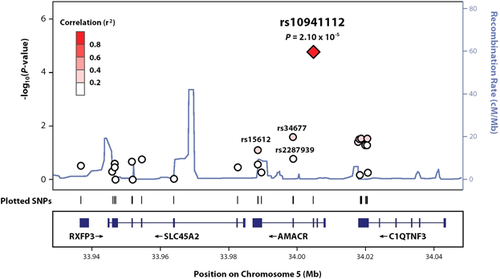
AMACR (UniProtKB Accession #: Q9UHK6) codes a key enzyme in the metabolism of fatty acids and bile acids derived from cholesterol, and has been previously implicated in schizophrenia (Bespalova et al., 2010), in particular the SNPs rs2278008 (p = 0.51) and rs2287939 (p = 0.13). To further characterize the association signal at rs10941112, we phased the haplotype structure of rs10941112, rs2278008, and rs2287939, as well as the missense variant rs34677 in exon 4 (G556T; V186F; MAF = 0.17), whose minor allele exhibits a significant protective effect on schizophrenia risk (β = −0.73 ± 0.63; p = 0.021) and was excluded from the 11,878 protein-altering variants selected for analysis because SIFT and PolyPhen2 scores were not assigned by ANNOVAR (note: SIFT score from the Variant Effect Predictor tool on the Ensembl website predicts damaging effects). In total, nine haplotypes were identified, with a significant association with schizophrenia observed for the major haplotype (f = 0.57; p = 0.0036; β = −0.73 ± 0.63), which harbors the rs10941112 risk allele (supplementary Table S1; Abecasis, Cherny, Cookson, & Cardon, 2002).
The next best signal is for the common SNP rs10378 (p = 2.80 × 10−5; MAF = 0.18) in the transmembrane protein gene TMEM176A (supplementary Figure S4; UniProtKB Accession #: Q96HP8), which also yielded a q-value of 0.073, with increased risk for the minor T allele (β = 1.14 ± 0.53), with a CADD score in the top 0.25% (G561T; L187F). Among affected individuals, a ratio of 3:15:7 is observed for genotypes TT: TG: GG, corresponding to an inflated frequency of 0.42 for the risk allele relative to the 0.15 estimated for European populations (1000 Genomes); whereas a skewed ratio of 0:31:78 was found for unaffected members, with a T allele frequency of just 0.14. Of the other SNPs presented in Table 2, all have q-values of 0.15 or greater, with no prior evidence implicating them in the disorder. At a permissive p-value of less than 0.01 (n = 118 SNPs; supplementary Table S2), no variants were found to map to any of the well-known schizophrenia candidate risk genes (e.g., DISC1, ZNF804A, NRG1, the MHC locus) (Harrison, 2015).
In an effort to replicate the significant associations at rs10941112 and rs10378, as well as investigate three other compelling missense variants within the gene AMACR (rs2278008, rs2287939, and rs34677), independent MGI samples were genotyped (n = 456). No significant associations were observed in these data, including rs10941112 (p = 0.79; β = −0.022 ± 0.16) and rs10378 (p = 0.92; β = 0.0015 ± 0.031), with the results found in supplementary Table S3. Additionally, we examined the PGC GWAS data for schizophrenia and similarly found no evidence for association at rs10941112 (p = 0.80) and rs10378 (p = 0.67).
3.3 Association results for PCET
Given the deficits in abstraction and mental flexibility observed in the study samples (see, section 2 for selection criteria), we also tested the rs10941112 and rs10378 for association with PCET efficiency scores (n = 105). Both show near significant associations with the neurocognitive measure (p = 0.087 and 0.053, respectively), with their estimated directions of effect consistent with heightened risk of schizophrenia (β = −0.26 ± 0.29 and −0.41 ± 0.41, respectively). Not surprisingly, bivariate analyses of schizophrenia and PCET efficiency revealed highly significant associations at both SNPs (p = 5.1 × 10−5 and 1.81 × 10−4, respectively).
3.4 mRNA transcript analysis
For the two significant association signals, rs10941112 and rs10378, we tested for cis effects on mRNA expression in lymphoblastoid cell lines. We detected a highly significant association between rs10941112 and the primary transcript of AMACR (RefSeq ID: NM_014324), with carriers of the disease risk allele showing increased expression levels (β = 0.18 ± 0.10; p = 5.53 × 10−4), and transcript levels showing association with schizophrenia in our MGI families (β = 1.08 ± 0.73; p = 3.95 × 10−3). Three other AMACR missense SNPs were also examined (supplementary Table S4), with rs34677 showing a significant association with expression (p = 6.84 × 10−3). As for rs10378, no significant effect was detected for TMEM176A transcript levels (NM_018487).
Interrogating data from the GTEx project, rs10941112 shows significant association with AMACR expression in various tissues, with the strongest signal observed for muscle-skeletal (n = 143; p = 2.7 × 10−18). For GTEx data from 12 different brain regions, significant associations were detected for seven, including the frontal cortex (n = 27; p = 0.03; supplementary Figure S5), a region highly implicated in schizophrenia pathology, with the nucleus accumbens yielding the most significant result (n = 30; p = 6.0 × 10−5). In each case, homozygous risk allele carriers showed increased expression of AMACR, consistent with our findings from the MGI families.
3.5 Pathway enrichment tests
To search for wider biological patterns in the data, pathway enrichment analyses were performed on gene-based association results. Genes with p-values <0.01 for the GATES association test were selected (n = 87), with AMACR (p = 2.10 × 10−5; Benjamini–Hochberg adjusted p = 0.072) and TMEM176A (p = 1.40 × 10−4) representing the top two association results for schizophrenia (Supplementary Table S5). Fisher's exact tests were conducted on these results, as well as randomized gene lists (100,000×), with the top five enriched pathways all related to bile acid biosynthesis and peroxisomal lipid metabolism (Supplementary Table S6; empirical p-values range from 0.0022 to 0.017; none significant after Bonferroni multiple testing correction), reflecting the presence of AMACR and HSD17B4 (GATES p = 6.24 × 10−3) in our gene list, both encoding enzymes involved in these processes.
We also examined pathway enrichment among PPI pairs, under the hypothesis that causal genetic variation affects a limited set of etiological mechanisms underlying schizophrenia, which are detectable by PPIs. Using the online database STRING v. 10.0, PPIs were identified for genes among our top GATES association results (nominal p < 0.05; n = 359 genes). The nodes (i.e., observed genes) of the resulting PPI network (supplementary Figure S6) were then tested (n = 47), revealing significant enrichment after Bonferroni correction in the Reactome pathway “NCAM Signaling for Neurite Outgrowth” (empirical p = 3.0 × 10−5), as presented in Table 3.
| Enriched pathway | Source | No. observed (%) | Empirical p-value | Observed genes |
|---|---|---|---|---|
| NCAM signaling for neurite outgrowth | Reactome | 4 (6.25) | 3.0 × 10−5* | COL2A1, COL4A2, COL9A1, SPTAN1 |
| Developmental biology | Reactome | 6 (1.51) | 1.3 × 10−4 | COL2A1, COL4A2, COL9A1, SPTAN1, PPARGC1A, TGS1 |
| NCAM1 interactions | Reactome | 3 (7.69) | 2.3 × 10−4 | COL2A1, COL4A2, COL9A1 |
| JAK-STAT signaling pathway | KEGG | 4 (2.58) | 3.4 × 10−4 | STAT2, IL11RA, IL11, IFNAR1 |
| RNA polymerase I promoter opening | Reactome | 2 (3.23) | 4.4 × 10−4 | HIST1H3A, HIST1H4A |
| Alpha-V beta-3 integrin pathway | PID | 3 (4.00) | 6.9 × 10−4 | COL2A1, COL9A1, ILK |
- Table presents enriched pathways (empirical p < 1.0 × 10−3) in PPI pairs and networks (n = 47 nodes) derived for a set of 359 genes representing the top associations for schizophrenia (GATES p < 0.05). PPIs were determined using the online database STRING v. 10.0 (experimental data only, with confidence scores greater than 0.40). Fisher's exact tests were permuted 100,000 times for each pathway (n = 1,273), with randomized gene lists drawn from the 7,300 genes tested for association via the GATES method. The four variables in the 2 × 2 contingency tables are as follows: (1) number of PPI genes in target pathway; (2) number of PPI genes not in target pathway; (3) number of genes in target pathway not among PPI genes; and (4) number of genes examined via GATES, excluding genes representing (2) and (3). The sources of the pathways are KEGG, Biocarta, Pathway Interaction Database (PID), and Reactome.
- * Bonferroni adjusted p = 0.038.
Lastly, we tested rs10941112 and rs10378 for pairwise interaction effects on schizophrenia with all the other exome variants examined in this study, revealing no significant effects (top p = 0.12). However, when the top 500 SNP-SNP interactions were analyzed for pathway enrichment, key brain-related pathways emerged for rs10941112 (Table 4), most notably mitogen-activated protein kinase (MAPK) signaling (empirical p = 2.0 × 10−4). For rs10738, regulation of the eIF-4 complex and p70-S6 kinase produced the strongest evidence of pathway enrichment (empirical p = 1.1 × 10−4; supplementary Table S7).
| Enriched Pathway | Source | No. observed (%) | Empirical p-value | Observed genes |
|---|---|---|---|---|
| MAPK signaling | KEGG | 10 (19.2) | 2.0 × 10−4* | MYC, NFATC4, BDNF, ECSIT, MAP2K6, EGF, CACNG6, CACNB2, CACNA1S, NTRK1 |
| TRAF6 sediated snduction of NFKB and Map Kinases | Reactome | 4 (30.8) | 2.6 × 10−3 | NOD1, IRAK4, ECSIT, MAP2K6 |
| Innate immune system | Reactome | 14 (17.1) | 3.7 × 10−3 | NOD1, DEFB129, DAK, DEFB132, ART1, P2RX7, IRAK4, ECSIT, MAP2K6, C8A, C9, TLR10, CASP8, NLRC5 |
| Agrin in postsynaptic differentiation | Biocarta | 5 (41.7) | 4.1 × 10−3 | NRG1, RAPSN, LAMA2, UTRN, PAK6 |
| Interleukin-1 signaling | Reactome | 3 (30.0) | 8.9 × 10−3 | NOD1, IRAK4, MAP2K6 |
- Table presents enriched pathways (empirical p < 0.01) for the top 500 pairwise SNP-SNP interaction effects on schizophrenia for rs10941112 (MGA model in SOLAR), with gene annotation determined with the program ANNOVAR. Fisher's exact tests were permuted 100,000 times for each pathway (n = 1,141), with randomized gene lists drawn from the 6,396 genes mapped to the 11,878 missense variants, weighted by SNP coverage. The four variables in the 2 × 2 contingency tables are as follows: (1) number of observed genes (i.e., those harboring SNP-SNP interaction effects, as defined above) in target pathway; (2) number of observed genes not in target pathway; (3) number of genes in target pathway not among observed genes; and (4) number of genes mapped to the 11,878 variants tested for interaction effects, excluding genes representing (2) and (3). The sources of the pathways are KEGG, Biocarta, Pathway Interaction Database (PID), and Reactome.
- * Bonferroni adjusted p = 0.23.
4 DISCUSSION
From whole exome sequences of 136 individuals from eight multiplex families, we have identified two protein-altering variants that are associated with schizophrenia: rs10941112 in the gene AMACR and rs10378 in TMEM176A. The product of AMACR, alpha-methyl-CoA racemase, plays a pivotal role in lipid and bile acid metabolism. AMACR deficiency (MIM 604489) can lead to the toxic accumulation of metabolic intermediates in tissues and blood plasma, most notably pristanic acid via the α-oxidation pathway, characterized by adult onset of neurodegenerative symptoms, including sensory motor neuropathy, pigmentary retinopathy, seizures, and neurocognitive impairment (Thompson et al., 2007). In one documented patient, AMACR deficiency was comorbid with schizophrenia, displaying elevated plasma pristanic acid levels and neurologic deficits resembling stroke-like episodes (Kapina et al., 2010). Molecular analysis of AMACR identified a homozygous mutation G559A (G187R) in the patient, which was absent in more than 100 tested controls. Interestingly, in a separate study, sequence analysis of a trio of AMACR deficiency patients showed two of the cases as homozygous carriers of the common rs10941112 mutation, hinting at its potentially damaging effects on enzymatic function (Ferdinandusse et al., 2000).
Although rs10941112 is significantly associated with AMACR transcript levels in both our families (p = 5.53 × 10−4) and GTEx tissue samples (smallest p = 2.7 × 10−18), we hypothesize that the missense variant does not directly regulate gene expression, but could reflect a feedback mechanism, whereby increased mRNA levels are triggered through unmetabolized pristanic acid due to deficient AMACR function, leading to neurodegenerative consequences and, in some cases, heightened risk for schizophrenia. Studies on the neurotoxic effects of pristanic acid on rat brain have revealed dramatic Ca2+ dysregulation, impaired aerobic metabolism, and neural cell death (Busanello et al., 2011). Moreover, alterations in white matter have been detected in cases of AMACR deficiency (Clarke et al., 2004), which, along with deficits in working memory and risk for retinitis pigmentosa, are features shared with schizophrenia (Du et al., 2013; McDonald, Kenna, & Larkin, 1998).
To date, a number of genetic studies of schizophrenia have detected positive linkage signals and structural changes at locus 5p13, the genomic location of AMACR (Bespalova et al., 2005; Cooper-Casey et al., 2005). Based on these findings, as well as the overlapping symptomatology of AMACR deficiency, Bespalova et al. (2010) identified AMACR as a candidate for schizophrenia susceptibility, screening its coding and flanking regions in a multiplex Puerto Rican pedigree. A significant association was observed for a phased haplotype of rs10941112 and two downstream missense variants, rs2287939 and rs2278008 from exons 4 and 5 (p = 3.6 × 10−5), with increased susceptibility among male carriers. Based on our exome sequence data, the LD relationship between rs10941112 and other nearby AMACR variants is relatively weak (Figure 1), with the strongest correlation observed for rs34677 (r2 = 0.36), a missense variant in exon 4 that shows modest evidence of association with schizophrenia (p = 0.021). To delineate the signal, we phased the haplotype structure of rs10941112 and rs34677, as well as the missense SNPs implicated by Bespalova et al. (2005) revealing a significant association between schizophrenia and the major haplotype harboring the rs10941112 risk allele (corrected p = 0.032).
However, despite this compelling evidence, we were not able to replicate the association signal at rs10941112. GWAS results from the PGC for a large case-control schizophrenia cohort show no significant association at this locus, nor genotypes that were generated for independent MGI samples (n = 456) for rs10941112 (p = 0.79) and other promising SNPs within AMACR (i.e., rs34677, rs2278008, and rs2287939). If rs10941112 is indeed a risk variant for schizophrenia, its incomplete penetrance is likely to be influenced by other genetic factors and their dynamic interactions within broader networks, with its effects potentially modulated by the polygenic burdens within individual families. Although clearly underpowered, we probed this hypothesis by testing rs10941112 for pairwise interactions with all the other exome variants on affection status, revealing no significant effects (top p = 0.12). However, when the top interactions were then tested for pathway enrichment, some key brain-related pathways emerged, most notably the top result, MAPK signaling (empirical p = 2.0 × 10−4). This highly conserved signaling cascade is involved in cell proliferation, differentiation and migration, including neuronal differentiation and control of synaptic plasticity (Thomas & Huganir, 2004), with some evidence for its role in the pathophysiology of schizophrenia (Funk, McCullumsmith, Haroutunian, & Meador-Woodruff, 2012). Interestingly, both lipids and bile acids appear to regulate parts of the pathway, including the subcellular localization of the MAPK signaling proteins, as well as their kinase activities (Anderson, 2006; Anwer, 2012).
It should also be noted that the MGI samples sequenced in this study were selected, in part, for their extremity in neurocognitive measures, in particular the PCET, which is compromised among individuals with schizophrenia and their relatives (Kos et al., 2016). This is borne out in the residuals generated for PCET efficiency scores under the polygenic model in SOLAR, as the mean for the exome sequence data (−0.29, SD = 1.12) is significantly different from the mean for the genotyped samples (−0.056, SD = 0.99; one-tailed p = 0.019). Interestingly, for our top two results, rs10941112 and rs10378, near significant associations were observed for PCET efficiency in the exome data (p = 0.087 and 0.053, respectively), but not in the follow-up genotype data (p = 0.71 and 0.96). This was also the case for bivariate analysis of PCET and affection status, with significant associations observed for these two variants in the exome data (p = 5.1 × 10−5 and 1.81 × 10−4), with a highly significant negative correlation (r = −0.38; p < 2.2 × 10−16) observed between the estimated beta coefficients for the two phenotypes for all 11,878 missense SNPs. This potential neurocognitive dimension of the observed risk effects in our data remains to be further explored, and may account for the non-replication of rs10941112 and rs10378 in schizophrenia cohorts not selected in this manner, in addition to the limited power of our study design. Moreover, lipidomic profiling of pristanic acid and related long-chain fatty acids in the MGI families and other schizophrenia cohorts will be necessary to establish the etiological link between AMACR enzymatic dysfunction and schizophrenia risk, as proposed here.
As for the association signal at rs10378, this is a novel finding with less supporting evidence in the existing literature. The missense variant is located in exon 5 of TMEM176A (supplementary Figure S4), a gene that encodes a transmembrane protein whose function remains largely unclear, although may have a role in the immune system (Condamine et al., 2010). TMEM176A has been implicated in animal models of depression, where a 35-fold difference in expression levels in cortex and hippocampus was observed between depression-sensitive and depression-resistant rat lines (Blaveri et al., 2010). Carriers of the alternate T allele (transversion G561T; amino acid substitution L187F) in MGI show increased susceptibility for schizophrenia, however, like rs10941112, this was not replicated in either the PGC GWAS results or the independent MGI genotype data (p = 0.93). The local LD structure for rs10378 is mostly negligible, with the exception of a lone correlation peak (r2 = 0.38) at intronic rs45455308 (p = 5.0 × 10−3 for schizophrenia risk) in the neighboring gene ABP1, which, notably, codes for an actin-binding protein that controls dendritic spine morphology and influences synapse formation (Haeckel, Ahuja, Gundelfinger, Qualmann, & Kessels, 2008). Pairwise interaction effects on schizophrenia were again examined, but did not yield any significant effects. However, pathway analysis of the top results did reveal strong enrichment of genes involved in the regulation of the eIF-4 complex and p70-S6 kinase (empirical p = 1.1 × 10−4), which plays a critical role in mRNA translation and synaptic plasticity (Klann, Antion, Banko, & Hou, 2004), with the phosphorylation cascade activated by various kinases, including ERK, p38 MAPK, and mTOR (Hoeffer & Klann, 2010; Thomas & Huganir, 2004).
The link to synaptic plasticity in our data is further underscored by our analysis of PPI pairs and networks, revealing significant enrichment of genes involved in neural cell adhesion molecule (NCAM) signaling for neurite outgrowth. NCAM is a transmembrane glycoprotein of the immunoglobulin superfamily and plays an important role in neural differentiation, as well as synaptic plasticity, learning, and memory (Senkov, Tikhobrazova, & Dityatev, 2012). Abnormalities in NCAM expression have been frequently linked to major psychiatric disorders, including schizophrenia (Piras et al., 2015).
5 CONCLUSION
From our analysis of whole exome sequence data, we report on two protein-altering SNPs, rs10941112 and rs10378 from the genes AMACR and TMEM176A, which are significantly associated with schizophrenia risk in multiplex families, with the former variant also displaying significant associations with gene expression. AMACR, which has been previously implicated in schizophrenia, encodes a pivotal enzyme in peroxisomal lipid oxidation, with deficiencies leading to neurotoxic accumulation of metabolic intermediates, which have been linked to a variety of neurological problems that manifest in adulthood, including neurocognitive impairment. Pathway analyses of PPI networks and SNP-SNP interaction effects reveal enrichment of genes involved in various pathways related to synaptic plasticity, in particular, kinase phosphorylation cascades known to affect learning and memory formation, with dysfunction in such pathways implicated in a number of psychiatric disorders. When tested against a measure of mental flexibility, both SNPs show near significant associations, indicating a neurocognitive dimension to their putative risk effects on schizophrenia, which was affirmed through bivariate analyses. Recognizing that functional improvement in schizophrenia patients will likely require treatment of cognitive deficits, these findings provide some insight into the potential etiological mechanisms involved in synaptic plasticity, as well as identify a promising lipid biomarker, pristanic acid, for enhanced diagnosis and therapeutic intervention that could complement current pharmacological strategies that largely target neurotransmitter dysfunction.
ACKNOWLEDGMENTS
The MGI study is supported by NIH grants MH061622, MH042191, and MH063480. Exome sequencing was funded in part by a Texas Biomedical Research Institute Forum Grant. Development of SOLAR was supported by NIH grant MH059490. This research was conducted in part in facilities constructed with support from Research Facilities Improvement Program Grant Number C06 RR017515 from the National Center for Research Resources, National Institutes of Health. We thank the participants of the MGI study, as well as our research staffs.
CONFLICTS OF INTEREST
We have no conflicts of interest, financial, or otherwise, to report.