Intragenic rearrangements in NRXN1 in three families with autism spectrum disorder, developmental delay, and speech delay†‡
The identified rearrangements have been deposited in GenBank: NRXN1_intragenic_deletion FJ972627; NRXN1_intragenic_duplication1 FJ972628; NRXN1_intragenic_duplication2 FJ972629.
How to cite this article: Wiśniowiecka-Kowalnik B, Nesteruk M, Peters SU, Xia Z, Cooper ML, Savage S, Amato RS, Bader P, Browning MF, Haun CL, Duda AW III, Cheung SW, Stankiewicz P. 2010. Intragenic Rearrangements in NRXN1 in Three Families With Autism Spectrum Disorder, Developmental Delay, and Speech Delay. Am J Med Genet Part B 153B:983–993.
Abstract
NRXN1 is highly expressed in brain and has been shown recently to be associated with ASD, schizophrenia, cognitive and behavioral abnormalities, and alcohol and nicotine dependence. We present three families, in whom we identified intragenic rearrangements within NRXN1 using a clinical targeted oligonucleotide array CGH. An ∼380 kb deletion was identified in a woman with Asperger syndrome, anxiety, and depression and in all four of her children affected with autism, anxiety, developmental delay, and speech delay but not in an unaffected child. An ∼180 kb tandem duplication was found in a patient with autistic disorder and cognitive delays, and in his mother and younger brother who have speech delay. An ∼330 kb tandem duplication was identified in a patient with autistic features. As predicted by conceptual translation, all three genomic rearrangements led to the premature truncation of NRXN1. Our data support previous observations that NRXN1 may be pathogenic in a wide variety of psychiatric diseases, including autism spectrum disorder, global developmental delay, anxiety, and depression. © 2010 Wiley-Liss, Inc.
INTRODUCTION
The genetic cause of autistic spectrum disorder (ASD) is recognized only in about 10–20% of cases. The most common cytogenetic abnormalities are maternally derived duplication of chromosome 15q11q13 found in 1–3% of cases [Cook et al., 1997; Abrahams and Geschwind, 2008; Marshall et al., 2008], deletions of 7q22q31, 22q13.3, 2q37, 18q21q23, and Xp22, duplications of 7q11.23 and 17p11.2, and aneuploidy of chromosome Y [Schaefer and Mendelsohn, 2008]. In addition, mutations in the FMR1, MECP2, TSC1, NF1, and PTEN genes that result in monogenic disorders: fragile X syndrome, Rett syndrome, tuberous sclerosis, neurofibromatosis, and PTEN hamartoma tumor syndrome, respectively, have been reported in patients with ASD [Muhle et al., 2004; Abrahams and Geschwind, 2008; Lintas and Persico, 2008; Morrow et al., 2008]. Other genes, for example, CNTNAP2, SHANK3, NLGN3, and NLGN4 that play an important role in the neuronal activity, have also been shown to contribute to the etiology of ASD [Abrahams and Geschwind, 2008; Lintas and Persico, 2008; Marshall et al., 2008; Morrow et al., 2008].
Recently, several studies have shown the importance of copy-number variations (CNVs) in the etiology of ASDs. Sebat et al. 2007 have reported de novo CNVs in 10% of simplex families with autism and in 2% of patients with an affected first-degree relative. Similar results have been obtained by Marshall et al. 2008, who identified de novo CNVs in ∼7% of patients with sporadic ASD and in ∼2% of families with affected siblings. Two or four de novo CNVs were found in ∼7.5% and ∼3.5% of these individuals, respectively. Christian et al. 2008 have identified CNVs in 11.6% patients with ASD (14% de novo and 86% inherited), Morrow et al. 2008 have found submicroscopic de novo CNVs in over 10% of patients with autism and Weiss et al. 2008 and Kumar et al. 2008 have reported common recurrent ∼600 kb reciprocal deletions and duplication in 16p11.2 in ∼1% of patients with autism. Moreover, Glessner et al. 2009 examined patients with ASD and found CNVs that belong to the ubiquitin gene family and genes that are implicated in neuronal development in 1.4% and 6.1% patients, respectively.
Some studies have suggested that genes influencing synaptic activity may contribute to the etiology of ASD [Marshall et al., 2008; Morrow et al., 2008; Glessner et al., 2009]. One of these gene families are neurexins (NRXN1, NRXN2, and NRXN3), transmembrane proteins and cell-surface receptors or cell-adhesion molecules in the nervous system that bind to postsynaptic cell-adhesion molecules: neuroligins, neurexophilins, and dystroglycans [Kirov et al., 2008; Nussbaum et al., 2008] and are essential in learning, memory, and cognition processes.
The function of neurexins in synaptic activity and the results of previous association studies make the neurexin genes strong candidates that contribute to ASD [Nussbaum et al., 2008]. However, mutations in neurexins have been found only in a small percentage of ASD patients [Feng et al., 2006; Autism Genome Project Consortium, 2007; Kim et al., 2008; Marshall et al., 2008; Zahir et al., 2008; Glessner et al., 2009]. Interestingly, a number of genomic rearrangements involving NRXN1 have been reported. Friedman et al. 2006 and Zahir et al. 2008 described a de novo ∼320 kb deletion in 2p16.3 identified with an Affymetrix GeneChip Human Mapping 100 K arrays that removed exons 1–5 in α-NRXN1 in a 7-year-old boy with cognitive impairment, autistic features, vertebral anomalies, and mild facial dysmorphism. The Autism Genome Project Consortium 2007 examined 1,181 families with at least two members affected with ASDs using Affymetrix 10 K SNP arrays. They identified a de novo hemizygous deletion of coding exons 6–18 in α-NRXN1 and exon 1 in β-NRXN1 in two siblings with typical autism and language regression. Kim et al. 2008 found two balanced chromosomal translocations with breakpoints in 2p16.3 in individuals with ASD; an insertional translocation ins(16;2)(q22.1;p16.1p16.3) removed exons 1–5 of α-NRXN1 and was inherited from the apparently normal father, whereas t(1;2)(q31.3;p16.3)dn had the 2p16.3 breakpoint mapping ∼750 kb 5′ to α-NRXN1. Genomic rearrangement involving NRXN1 have also been reported in patients with schizophrenia. Kirov et al. 2008 examined 93 individuals affected with schizophrenia using tiling path BAC array CGH and in two affected siblings found one ∼250 kb deletion disrupting α-NRXN1 inherited from their unaffected mother. Walsh et al. 2008 reported a 115 kb deletion disrupting NRXN1 in a patient with childhood-onset schizophrenia and recently, Rujescu et al. 2009 examined almost 3,000 patients with schizophrenia and found 14 (0.47%) rearrangements in NRXN1, including de novo CNVs, and in 0.15% of controls. The association became more significant when deletions and duplications that disrupted exons were analyzed (CNVs in 0.17% of patients and in 0.020% of controls). They also found NRXN1 deletions in five patients with autism, alcohol dependence and a first-degree relative of a schizophrenia patient. This study has statistically demonstrated that exon-disrupting rearrangements in NRXN1 are more likely to be pathogenic.
We present three families in whom we identified an intragenic deletion and duplication within NRXN1 in patients with ASD, anxiety and depression, developmental delay, and speech delay.
CLINICAL REPORT
Family 1
Pedigree of Family 1 is shown in Figure 4d and clinical features are summarized in Table I.
| I-2 | II-1 | II-2 | II-3 | II-4 | II-5 | |
|---|---|---|---|---|---|---|
| Pyschological testing | + | + | + | − | GARS-2 | − |
| Height | 75–97% | 75% | 50–75% | 95% | 50% | 50% |
| Weight | 25–50% | 75–97% | 25% | 90–95% | 50–75% | 50% |
| Head circumference | 50% | 97% | 50% | 50–75% | 50% | 50% |
| Inner canthal distance | 75% | 50–75% | 50% | 97% | 75% | 75–97% |
| Outer canthal distance | 50–75% | 50–75% | 3–25% | 50–75% | 25–50% | 25% |
| Palpebral fissures | −3 SD | −4 SD | −6 SD | −2 SD | −5 SD | +1 SD |
| Nose | Prominent | Prominent | Prominent | N | Bulbous | Bulbous |
| Philtrum | 25% | 50–75% Mildly flat | 50–75% Mildly flat | 50% | 3–25% | 50% |
| Ear | 50-75% | 75% | 25% | 3–25% | 75–97% | 25% |
| Mouth | Wide | Full lips | Wide | N | Lower lip frenulum | N |
| Chest | N | >97% | 75% | 75–97% | Pectus 75% excavatum | 75% |
| Heart | N | 2/6 murmur | N | N | 1/6 murmur | N |
| Abdomen/back | N / N | N/2 Café au Lait | N/N | N | N/N | N/N |
| Hand | 50% | 75–97% | 50% | 75% | 50–75% | 25% |
| Third finger | 50% | 25% | 50% Pointed finger fat pads | 25–50% | 25% Pointed finger fat pads | 25% |
| Foot | 75–97% | 75–97% | 3–25% | 75% | 50–75% | <3% |
| Neurological/deep tendon reflexes | 2+ | 1+, poor balance, hypotonia | 2+,/ 3+, poor balance | 2+ | +1 | +2/+3 |
| Creases/other findings | Hockey stick creases, R Variant Simian crease | Sydney lines bilateral | N | N | Hockey stick, Branchial sinus remnant | N/Cutis marmorata |
- N, no abnormality found.
I-2
The patient is a 36-year-old female, who was seen because of concerns regarding her 12-week pregnancy. She had a history of 10 pregnancies with four live-born children and five first-trimester spontaneous abortions. All of her children are full siblings. In addition, she had a history of anxiety, depression, and Asperger syndrome. A recent psychological exam found her to have low average cognitive ability, marginal memory, inattention, distractibility, and problematic reality testing. Completion of the ADI-R assessment confirmed a diagnosis of ASD. In the past, she has been hospitalized three times for psychosis, anxiety, and depression and has been treated with antipsychotic and antianxiety medications. Her first psychiatric hospitalization occurred at age 12 because of anorexia. Currently, she is managed as an outpatient. She was born prematurely at 32 weeks gestation and her birth weight was 1.36 kg (10 percentile). She was transferred to the Neonatal Intensive Care Unit and required oxygen supplementation. She later received speech therapy, attended regular school, but had trouble reading and focusing.
On physical exam, she appeared younger than her stated age. The patient exhibited some dysmorphic features, including a small thin face, with a high forehead, broad eyebrows, small palpebral fissures, mild epicanthal folds, prominent nasal root and bridge, wide nostrils, wide mouth, a narrow pointed chin, and prominent tragus on both ears (Fig. 1).
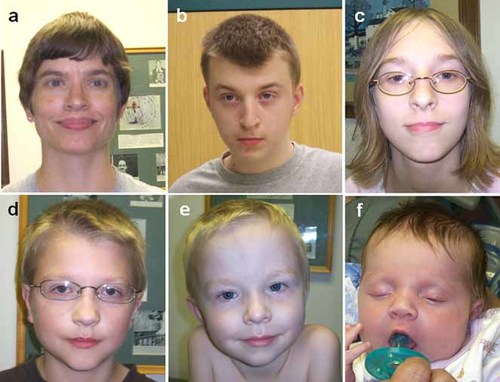
a: Proband I-2 in Family 1 at the age of 36 years, b; her first child at the age of 14, c: second child at the age of 12, d: third child at the age of nine, e: fourth child at the age of four, and f: fifth child at the age of 1 month. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
II-1
Family history was significant for the proband's oldest child, a 14-year-old male, is maintained in a residential hospital because of ASD. He had normal growth parameters at birth and no perinatal complications were reported. He showed developmental delay and an infant stimulation program was started. He had frequent otitis media and a single seizure episode at 1 month of age for which he was placed on Phenobarbital. He crawled at 9 months, walked at 15 months but attained few words by age 2 years. At 4 years of age, he was unusually frightened by adults and could not attend preschool. His psychocognitive assessment at 5 years of age revealed findings consistent with ASD. He attended special education from kindergarten to grade 6. His speech was characterized by echolalia. He had an excellent memory, but poor self-help skills. At age 12, he was evaluated for residential placement and accepted at a state residential hospital. He was removed from his home because of problems with significant aggressive behavior. After a trial in a foster home, placement was pursued. Evaluation at the time of admission indicated an IQ of 38, Peabody Picture Vocabulary (IIIA) IQ equivalent of 40, Alpern Ball Developmental Profile II IQ equivalent of 40, Vineland Adaptive Behavior Score of 56. Completion of the ADI-R assessment determined a diagnosis of ASD.
Physical exam showed him to speak in three- to four-word sentences, to exhibit perseverative and repetitive speech. He had an oval face, high forehead with bitemporal recession of hairline, arched eyebrows, prominent nasal root and bridge, wide mouth with upper lip bow-shaped, narrow pointed chin, low posterior neck hair, a mild pectus excavatum, chest and inner nipple distances were greater than 97 percentile, an innocent murmur 2/6 at the left sternal border, joint hypermobility and soft skin were noted. Mild hypotonia and incoordination were noted. There were two café au lait spots on his back (3.5 and 2.5 cm in diameter) (Fig. 1).
II-2
The proband's second live-born child, a 12-year-old girl, was evaluated for symptoms of anxiety and poor attention span. She had increasing problems including sleepwalking, trichotillomania, shoplifting, aggressive behavior, head banging, inappropriate or incomplete understanding of social situations, and lack of empathy. She is the product of a normal gestation and delivery. Developmental milestones were reported as normal. She was in foster care between ages 4 and 5 because of neglect and witnessing domestic violence.
On physical exam, she appeared younger than her stated age but with normal growth parameters. She has an oval-shaped face with high forehead and wears glasses to correct myopia. She has arched eyebrows, prominent nasal root and bridge, protruding nose, mildly flattened philtrum, wide mouth, pointed chin as well as slight contractures of joints. Her neurological examination demonstrated mild balance problems on tandem walking. Neuropsychological testing showed full-scale IQ of 58. Verbal expression was characterized by poor vocabulary but adequate articulation. The Aphasia Screening Test indicated impaired language functions. Specifically, elements of dysnomia, spelling dyspraxia, dyslexia, dyscalculia, construction dyspraxia, left/right confusion were noted. The Vineland Adaptive Behavior Scales testing indicated impaired socialization skill (SS = 69; AD = 7 years, 2 months). Her overall adaptive behavior composite was in the mildly impaired range (SS = 6; AD = 8 years, 4 months). Completion of the ADI-R assessment determined that the patient fits a diagnosis of autism (Fig. 1).
II-3
The proband's third live-born child, a 9-year-old boy, was able to function well academically and socially. He was the product of a full-term pregnancy and spontaneous delivery with normal birth weight and length. His past medical history was significant for a single seizure (likely due to a febrile illness) at 11 months and the placement of PE tubes for recurrent otitis media.
On physical exam he exhibited an oval-shaped face and wore glasses for myopia. His hairline was characterized by an upsweep pattern. He had a broad nasal bridge, thin lips, and normal jaw. His ears showed a prominent anthelix, Darwinian tubercle, and a mole on the left. He had a mild pectus excavatum and a single café au lait spot on the abdomen. The neurological exam was normal (Fig. 1).
II-4
The proband's fourth live-born child, a 4-year-old boy, was born after a full-term pregnancy and a normal vaginal delivery. His motor developmental milestones were mildly delayed. The patient started saying single words at age 8 months but little progress had been achieved for the next 8–10 months. Because of his speech delay and autistic features, the patient was evaluated by the GARS-2 Rating Scale which revealed an Autism Index of 104 (61 percentile; Very Likely Probability of Autism). He required placement of PE tubes for frequent otitis media.
On physical exam, he had a prominent central forehead, arched eyebrows, strabismus of the left eye, and mild inner epicanthal folds. He had a broad nasal root and a bulbous nasal tip, delicate upper lip but wide mouth, and mild ankyloglossia. Ears showed prominent anthelix and rather flat helix. There was a branchial fistula remnant on the anterior midline of the neck. There was a mild pectus excavatum, mild winging of the scapulae, and a shawl scrotum. There is a hockey stick crease on the left palm. The neurological exam was unremarkable (Fig. 1).
II-5
The proband gave birth to a full-term infant girl. Birth weight was 3.5 kg (50–75 percentile) and length was 20.5 in. (75th percentile). There was a prominent forehead, arched eyebrows, and bi-temporal narrowing. The nose had a broad root and was upturned with a bulbous tip. There was a mild micrognathia and mildly pointed chin. The ears were somewhat cup-shaped with a prominent anthelix. There were extra skin folds on the neck. The neurological examination including neonatal reflexes was unremarkable (Fig. 1).
Family 2
Pedigree of Family 2 is shown in Figure 5c.
II-1
The proband was born at 36-week gestation by spontaneous vaginal delivery followed by an uncomplicated neonatal course. He had a history of feeding challenges, especially with chewy textures, and a diagnosis of gastroesophageal reflux disease. He underwent an initial developmental evaluation at age 3 years 4 months, at which time he was diagnosed with speech and language delay, and Pervasive Developmental Delay (PDD-NOS). At age 3 years 11 months, his performance on the Autism Diagnostic Observation Schedule (ADOS) placed the patient in the Autism Spectrum Disorder range. Completion of the Autism Diagnostic Interview—Revised (ADI-R) assessment determined a diagnosis of ASD. While he demonstrates delays in expressive language and language processing as well as cognitive delays, his physical growth was age-appropriate. He demonstrated a prominent forehead, arched eyebrows, broad upturned nasal tip, generous mouth, small and pointed chin. He was described as having sudden and unexplained mood changes (Fig. 2).
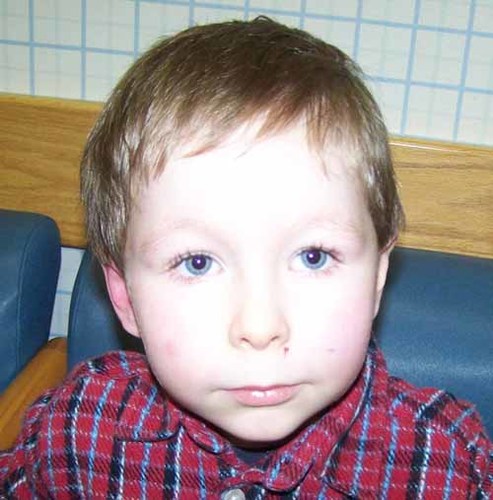
Proband II-1 in Family 2 at the age of 4 years. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
II-2
The patient's younger brother has not yet been formally evaluated. However, based on the observations made during the family's visit, he seemed to demonstrate a clinical presentation similar to the proband. At 18 months, this brother had no expressive language.
I-1
The 26-year-old mother of the proband had right-sided hearing loss and migraines. She reported having a history of speech problems and difficulty with social situations, but she had not had a psychological evaluation.
I-2
No abnormalities were reported in the proband's father.
Family 3
Pedigree of Family 3 is shown in Figure 6c.
II-2
The 7-year-old girl was first clinically diagnosed with ASD/Pervasive Developmental Disorder (PDD-NOS) at age 3. At age 7 years 5 months the ADOS and ADI-R were used to confirm her diagnosis. The patient met diagnostic cutoffs for Autism on the ADOS. On the ADI-R, she did not meet diagnostic criteria for either Autism or Autism Spectrum Disorder, however, the patient's mother was not a reliable informant, and that likely affected the outcome of the measure. The proband is the first live born child of 32-year-old mother and a 39-year-old father and was the product of an uncomplicated pregnancy. Her mother had a spontaneous abortion at 5 months gestation and describes some difficulty in school. The proband's height is at the 78th percentile and weight is at the 90th percentile. Her head circumference is 51.5 cm (80 percentile). On exam, she had a prominent forehead, bitemporal narrowing, prominent midface hypoplasia with decreased intercanthal bilateral distance, mild epicanthal folds, and a flat nasal bridge. Her slightly large ears demonstrated an unusual helix with decreased antihelix and smooth border with prominent helical curve. She displays a somewhat short neck, prominent gynecomastia with inverted nipples, and some mild truncal obesity. The patient has very small hands and fingers with puffy dorsum of the hand, and lower extremities show pes cavum with very prominent puffy dorsum of the foot and short toes. Motor, tone, and strength evaluation demonstrated slight truncal hypotonia. DTR's were normal. Developmentally, the patient continues to make progress in all areas; she has increased her vocabulary and social interaction (Fig. 3).
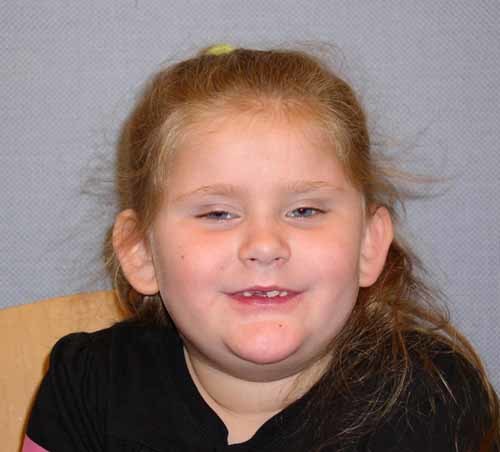
Proband II-2 in Family 3 at the age of 7 years. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Her mother (I-2) does report some atypical behaviors including intermitting mouthing of objects and food-hoarding. Karyotype and fragile X were normal. Her father (I-1) is dyslexic and has multiple psychiatric issues. The patient also has a 2½-year-old maternal half-brother (II-3), who has a speech delay but normal gross motor development and no other major medical issues.
METHODS
Blood samples from three probands (Patients I-2 in Family 1, II-1 in Family 2, and II-2 in Family 3) each suspected of having a subtle chromosome abnormality were submitted for testing at the Medical Genetics Laboratories (MGL) at Baylor College of Medicine (BCM) by the targeted oligonucleotide Chromosome Microarray Analysis (CMA) V6.3 OLIGO, V6.4 OLIGO, and V6.5 OLIGO. When the abnormalities were detected, family studies, clinical correlations, and genetic counseling were recommended. At the time of the family studies, medical records were requested and blood samples were obtained on each family member after informed consent using a protocol approved by the Institutional Review Board for Human Subject Research at Baylor College of Medicine. The Autism Diagnostic Observation Schedule (ADOS) and Autism Diagnostic Interview—Revised (ADI-R) were used in diagnostic evaluations of our patients.
Targeted Array CGH
Patients' genomic DNA was extracted from peripheral blood using Puregene DNA isolation kit (Gentra System, Inc., Minneapolis, MN). Initial targeted oligonucleotide array CGH was performed using CMA V6.3 OLIGO (Patients I-2 in Family 1), V6.5 OLIGO (Patient II-1 in Family 2), and V6.4 OLIGO (Patient II-2 in Family 3) designed by BCM MGL (http://www.bcm.edu/geneticlabs/cma/tables/44KDisorders.pdf) and manufactured by Agilent Technology (Santa Clara, CA). The array containing 42,640 BAC clone emulating oligonucleotides, with an average of 28 oligos corresponding to each BAC clone genomic locus was used as described [Ou et al., 2008]. The NRXN1 gene is represented by BAC clone RP11-800C7 emulating 33 oligonucleotides (chr2:50,128,256–50,295,947) that cover only 3′ portion of NRXN1 (chr2:49,999,148–51,113,178).
Whole-Genome Array CGH
Whole-genome oligonucleotide microarray CGH analysis was performed using NimbleGen array HG18_WG_CGH_v1 with 385,000 (Patients I-2 in Family 1 and II-1 in Family 2) and 2.1M (Patient II-2 in Family 3) oligonucleotides (NimbleGen Systems, Madison, WI) according to the manufacturer's instructions. The array containing 385,000 or 2.1 M variable length oligonucleotides (median probe spacing 6 or 1.5 kb) designed for unique sequence regions. The lengths of the oligonucleotides (50–85 mer) were designed so as to achieve equal melting temperature. Total genomic patient DNA was directly labeled with 9-prime-Cy5 mers and the reference DNA was labeled with 9-prime-Cy3. Probes were co-hybridized to slides using the MAUI Hybridization System (BioMicro Systems, Salt Lake City, UT). After posthybridization washing, the arrays were scanned using a GenePix 4000B Scanner (Molecular Devices Corporation, Sunnyvale, CA). Data were analyzed using NimbleScan v2.4 software and visualized with SignalMap (NimbleGen). The relative intensity of the test sample versus the reference DNA was indicated on a log2 scale. A positive result was determined when a genomic segment complementary to oligo probes for CNV (gain or loss) was 0.2-fold average difference from reference normal DNA.
FISH Analysis
Confirmatory FISH analysis in Families 1 and 3 were performed using standard procedures. Briefly, the BAC (bacterial artificial chromosome) clone RP11-800C7 was grown in TB media with 20 µg/ml chloramphenicol. DNA was extracted (Eppendorf Plasmid Mini Prep kit, Hamburg, Germany) and directly labeled with SpectrumOrangeTM dUTP by nick translation (Vysis, Downer Grove, IL) according to the manufacturers' instructions. Digital FISH images were captured by a Power Macintosh G3 System and MacProbe version 4.4 (Applied Imaging, San Jose, CA).
Long Range PCR and DNA Sequencing
LR-PCR reaction was performed to amplify the predicted junction fragments from the breakpoint regions according to manufacturer's instructions (Takara Bio, Inc., Shiga, Japan). PCR products were purified with the PCR Purification Kit (Qiagen, Valencia, CA) and sequenced by Sanger dideoxy sequencing (SeqWright, Lone Star, Houston, TX).
Bioinformatics and In Silico Sequence Analysis
Genomic and protein sequences defined based on the oligonucleotide coordinates from the array CGH experiment were downloaded from the UCSC genome browser (Build 36, UCSC genome browser, March 2006) and assembled using the Sequencher software (Gene Codes Corporation, Ann Arbor, MI). Interspersed repeat sequences were analyzed by RepeatMasker (http://genome.ucsc.edu).
RESULTS
Family 1
Array CGH
In Patient I-2, CMA V6.3 OLIGO revealed a loss of all 33 oligos corresponding to the BAC clone RP11-800C7 in the NRXN1 gene (Fig. 4a). Genome-wide oligonucleotide aCGH mapped the proximal breakpoint between 50,028,806 and 50,035,204 and the distal breakpoint between 50,412,598 and 50,418,802 (Fig. 4b), consistent with an ∼380 kb intragenic deletion. The other identified changes represented known copy-number polymorphisms.
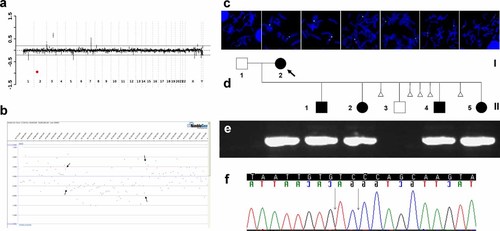
a: Targeted array CGH plot obtained with V6.5 oligo (Agilent 44 K) showing deletion in 2p16.3 in Family 1 (red mark). b: Whole genome oligo array CGH plot (NimbleGen 385 K) mapped the proximal breakpoint between 50,028,806 and 50,035,204 and the distal breakpoint between 50,412,598 and 50,418,802. c: Metaphase chromosomes after FISH with BAC clone RP11-800C7 in all Family 1 members studied. d: Pedigree of the Family 1. e: The 1.5% agarose gel with the 7251 bp PCR product that span the deletion junction fragment found in the proband and her four children. f: Chromatogram of the DNA sequence of the junction fragment. The vertical black arrow indicates the junction point.
FISH analysis
The array data were verified by FISH analysis with a BAC clone RP11-800C7 (Fig. 4c). Further FISH studies in I-1, II-1, II-2, II-3, II-4, and II-5 revealed that the deletion is also present in the proband's four children, II-1, II-2, II-4, and II-5 (Fig. 4c,d). One child II-3 had a normal result.
PCR and DNA sequencing
The 725 bp junction fragment of the deletion in Patient I-2 was amplified with use of LR-PCR with the following primers F: CCCCAAATAAGCCAGTATCTGAGTAGCATCT, R: GTAGAGGTTGTTGGTGTGATGGCCAGTATTA (Fig. 4e) and sequenced. Consistent with the FISH findings, the same junction fragment was found in the proband's four children (Fig. 4d,e). The proximal breakpoint was precisely mapped at 50,034,351 and the distal breakpoint at 50,413,346 bp (nucleotide numbering is based on human genome build 36, UCSC genome browser, March 2006). This 378,998 bp deletion removed exons 2–4 in. isoform β (exons 19–22 in isoform α), predicting by conceptual translations a premature truncation of NRXN1 (Figs. 7a and 8b,g). DNA sequence analysis of the breakpoint regions revealed 2 bp microhomology (Fig. 4f). Both breakpoints map within a unique DNA sequence.
Family 2
Array CGH
In patient II-1, CMA V6.5 OLIGO revealed a gain of 13 out of 33 oligos corresponding to the BAC clone RP11-800C7 in the NRXN1 gene (Fig. 5a); this gain was verified by genome-wide array CGH. The proximal breakpoint was mapped between 50,006,408 and 50,012,728 bp and the distal breakpoint between 50,193,772 and 50,200,141 bp (Fig. 5b). The other identified changes represented known copy-number polymorphisms.
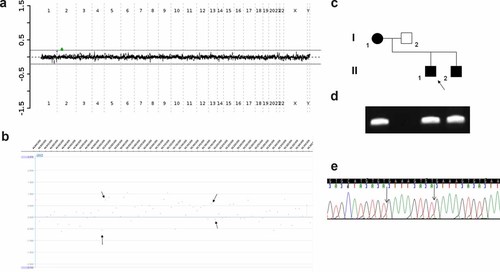
a: Targeted array CGH plot obtained with V6.5 oligo (Agilent 44 K) showing duplication in Family 2 (green mark). b: Whole genome oligo array CGH (NimbleGen 385 K) mapped the proximal breakpoint between 50,006,408 and 50,012,7281 bp and the distal breakpoint between 50,193,772 and 50,200,1411 bp. c: Pedigree of the Family 2. d: The 1.5% agarose gel with the 3671 bp PCR product that span the tandem duplication junction fragment found in the proband, his mother and brother. e: Chromatogram of the DNA sequence of the junction fragment. The vertical black arrow indicates the insertion GAAAGTGT.
PCR and DNA sequencing
The predicted tandem duplication junction fragment in Patient II-1 was amplified with use of LR-PCR and subsequently regular PCR with the following primers F: GTTGATGGTGGGGAGACTGT, R: GTGTTCAACCCCCTTGCTTA (Fig. 5d), and sequenced. The same 367 bp junction fragment was found in the proband's mother and younger brother (Fig. 5c,d). The proximal breakpoint was mapped 50,011,487 bp and the distal breakpoint at 50,195,746 bp, consistent with a 184,259 bp tandem duplication that copies exons 3–5 in isoform β (exons 20–23 in isoform α) and by conceptual translations predicted to result in a premature truncation of NRXN1 (Figs. 7a and 8c,h). DNA sequence analysis of the junction fragment revealed the presence of an eight nucleotide insertion: GAAAGTGT that match to sequence at the beginning of the duplication (Fig. 5e). Microhomology (2 bp) was found at the breakpoint regions. The proximal duplication breakpoint mapped within a unique sequence and the distal breakpoint mapped within a 1,1322 bp repetitive LINE/L1 element L1MC1.
Family 3
Array CGH
In patient II-2, CMA V6.4 OLIGO revealed a gain of all 33 oligos corresponding covering the 3′portion of the NRXN1 gene and a gain of 31 oligos emulating BAC clone RP11-109N23 on chromosome 14q24.2 (Fig. 6a). The 14q24 duplication was determined by FISH analysis to be inherited from the healthy mother and thus not likely clinically relevant. The duplication of NRXN1 on chromosome 2p16.3 was not found in the mother. The father and the other family members were not available for the studies.
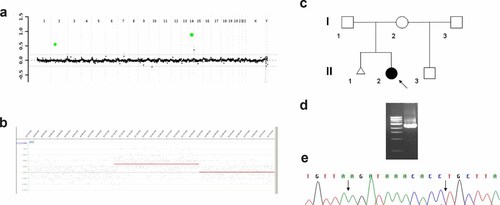
a: Targeted array CGH plot obtained with V6.4 oligo (Agilent 44K) showing gains in chromosomes 2p16.3 and 14q24.2 in Family 3 (green marks). b: Whole genome oligo array CGH (NimbleGen 2.1M) mapped the proximal breakpoint between 50,106,298-50,107,420 and the distal breakpoint between 50,435,668–50,437,268. c: Pedigree of the Family 3. d: The 1.5% agarose gel with the 2605 bp PCR product that span the tandem duplication junction fragment found in the proband. e: Chromatogram of the DNA sequence of the junction fragment. The vertical black arrow indicates the 11 bp insertion AGATAAACACC.
Genome-wide oligonucleotide aCGH narrowed the proximal breakpoint of the 2p16.3 duplication between 50,106,298–50,107,420 and the distal breakpoint between 50,435,668–50,437,268 (Fig. 6b). The 14q24.2 duplication extends between 72,232,861–72,234,122 and 72,695,291–72,696,109. The other identified changes represented known copy-number polymorphisms.
PCR and DNA sequencing
The predicted tandem duplication junction fragment was amplified with use of LR-PCR and subsequently regular PCR with the following primers F: CGGAGAACTGTGTGTTTATCTTGACAGGTG, and R: GTCTCCTTGTTAGATACCTAGGAAGAGTGGGTA (Fig. 6d), and sequenced. The proximal breakpoint was mapped at 50,106,413 and the distal breakpoint at 50,437,234 bp, consistent with a 330,821 bp tandem duplication that copies exons 1–4 in. isoform β and exons 19–22 in isoform α, and by conceptual translations predicted to result in the same premature truncation of both isoforms of NRXN1 (Figs. 7a and 8d,e,i). The truncated isofom β is predicted to be expressed in addition to the normal protein translated from the NRXN1 copy on the normal allele and the intact copy of NRXN1on chromosome dup(2)(p16.3p16.3).
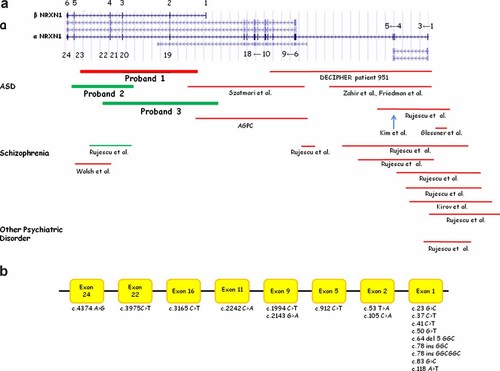
Schematic representation of a genomic structure of different isoforms of NRXN1 (UCSC genome browser, March 2006) showing exon locations for α-NRXN1 and β-NRXN1 and other isoforms predicted by RefSeq, UniProt, GenBank, CCDS, and Comparative Genomics. Summary of the reported a: genomic rearrangements in NRXN1 and b: mutations. The red bars indicate deletions, the green bars indicate duplication, and the blue arrow indicates insertion.
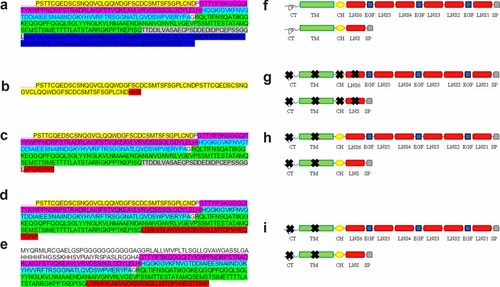
Amino acid sequence of a: 3′ portion of a normal isoform of α-NRXN1 protein, and the predicted structures of the 3′ portion of α-NRXN1 in b: Family 1, c: Family 2, and d: Family 3. Amino acid sequence of e: Predicted structure of the isoform of the β-NRXN1 protein in Family 3. f: Domains of the NRXN1 protein: LNS—laminin, neurexin, sex hormone-binding globulin, EGF—epidermal growth factor sequence, CH—carbohydrate region, TM—a transmembrane domain, CT—intracellular region. The predicted truncated NRXN1 in g: Family 1, h: Family 2, and i: Family 3. The colors designate the amino acid portions of NRXN1 encoded by different exons: yellow—exon 18 in isoform α, purple—exon 2 in isoform β and exon 19 in isoform α, light blue—exon 3 in isoform β and exon 20 in isoform α, green—exon 4 in isoform β and exon 21 in isoform α, gray—exon 5 in isoform β and exon 22 in isoform α, and dark blue—exon 6 in isoform β and exon 23 in isoform α. The non-highlighted aminoacids represent exon 1 of isoform β. Amino acids highlighted with red color depict the changed and truncated portion of the β-NRXN1 protein.
DNA sequence analysis of the junction fragment revealed the presence of an 11 nucleotide insertion: AGATAAACACC. Microhomology (1 bp) was found at the breakpoint regions. The distal breakpoint maps within a 1,591 bp MIRb (SINE/MIR) repetitive element and the proximal breakpoint maps in the unique sequence.
To determine whether the identified rearrangements are benign CNVs, we have screened our database of array CGH studies performed in over 9,000 patients analyzed by oligonucleotide CMA (Versions 6.2–7.4M). Of these patients, approximately 4,700 patients were tested on V6.2–V6.5 (44 K array) with 33 oligos, 431 patients V7.0 (105 K array) with 76 oligos covering the region chr2:50,008,520–51,105,061, and over 4,000 patients using V7.2–V7.4M (105 K array) with 130 oligos targeting all NRXN1 exons between chr2:50,001,386–51,109,503. In this series, there were predominantly children with developmental delay/mental retardation (18.5%), multiple congenital anomalies (14.6%), dysmorphic features (7.3%), autism or autistic spectrum (6.3%), seizures (4.4%), or others. We found only one additional case of intragenic deletion (max deleted region chr2:50,890,667–51,052,471), removing exons 4–5 of NRXN1 isoform alfa, and thus being potentially pathogenic. The indication for the CMA study was microcephaly. Unfortunately, this patient was unavailable for further clinical studies.
DISCUSSION
We describe three novel CNVs within NRXN1 in patients with a common phenotype similar to the phenotype of previously reported patients with a de novo deletion of NRXN1 [Zahir et al., 2008]. We believe that this clinically identifiable syndrome includes ASD, high anterior hairline, broad medial or arched eyebrows, strabismus, wide nasal bridge in the younger patients, prominent nasal root and bridge with slightly bulbous nasal tip in the older subjects, mild inner epicanthal folds, abbreviated or flat philtrum, wide mouth, mild ear anomalies, and pointed chin. A branchial sinus remnant was seen in one patient, and hearing loss in another.
The identified rearrangements in families 1–3 do not overlap with the vast majority of the reported mutations and CNVs in NRXN1 (Fig. 7). The deletion in Family 1 removed exons 2–4 in isoform β (exons 19–22 in isoform α), the duplication 1 in Family 2 copied exons 3-5 in isoform β (exons 20–23 in isoform α) and the duplication 2 in Family 3 copied exons 1–4 in isoform β (exons 19–22 in. isoform α). These CNVs are likely the result of non-homologous end joining [Roth and Wilson, 1986; Shaw and Lupski, 2004] or the recently described microhomology-mediated break-induced replication error (FoSTeS, MMBIR) mechanism [Slack et al., 2006; Lee et al., 2007; Payen et al., 2008], as eight and 11 nucleotide insertions GAAAGTGT and AGATAAACACC have been identified at the duplication junction fragment in Families 2 and 3, respectively (Figs. 5e and 6e) [Vissers et al., 2009].
The α-neurexin proteins contain three modules, each consisting of two LNS domains (laminin, neurexin, sex hormone-binding globulin) separated by an EGF-like (epidermal growth factor) sequence. These three repeats are followed by a carbohydrate region, a transmembrane domain and a short intracellular region (Fig. 8f). In the β-neurexins, a single LNS domain replaces the three repeats [Rowen et al., 2002; Kang et al., 2008; Yan et al., 2008; Zahir et al., 2008]. The deletion in Family 1 truncated the LNS6 domain (Fig. 8b,g) and the transmembrate domain in Families 2 and 3 (Fig. 8c–e,h,i).
The pathogenicity of the presented genomic rearrangements in Families 1–3 are supported by appropriate segregation of the CNVs with autistic phenotypes in the presented families, were absent in the unaffected family members, and have not been found in over 9,000 patients analyzed by oligonucleotide CMA. We did not study a matched control set of healthy children; however, we have not seen any CNVs involving NRXN1 in ∼200 parental studies. In addition, Glessner et al. 2009 examined more than 2,000 patients with ASD and found rearrangements in NRXN1 in ∼0.4% of patients but none in controls. Notably, all of these CNVs were inherited.
Furthermore, the predicted consequences of these genomic rearrangements are premature truncations. However, in Family 3, the truncated isofom β is predicted to be expressed in addition to the normal protein expressed from the intact copy of NRXN1 on the rearranged chromosome. Additionally, Rujescu et al. 2009 and Bucan et al. 2009 have demonstrated recently that exon disrupting rearrangements in NRXN1 are more likely to be pathogenic.
The phenotypic effects of abnormal NRXN1 are not clear and incomplete penetrance has been reported [Kim et al., 2008; Kirov et al., 2008]. The fact that some rearrangements in NRXN1 may occur in healthy individuals [Kim et al., 2008; Kirov et al., 2008; Rujescu et al., 2009] indicate that CNVs in the neurexin gene probably have various pathogenicity depending on the functional significance of different protein domains that are affected.
An increasing number of neurological disorders resulting from genomic rearrangements and CNVs have been described recently [Lee and Lupski, 2006; Gu and Lupski, 2008]. We believe that pathogenic (intra)genic and exonic CNVs are under-ascertained and suggest an application of exon targeted oligo array CGH for their efficient identification [Erez et al., 2009]. Our data confirm previous observations that NRXN1 may be pathogenic in a wide variety of psychiatric diseases, including autism spectrum disorder, anxiety, depression, ADHD, and developmental delays and suggest that such exon targeted arrays should be performed to identify pathogenic rearrangements in individuals presenting with these conditions.
Acknowledgements
We thank Dr. James R. Lupski, Dr. Marwan Shinawi, Dr. Joanna Wiszniewska, Dr. Heather Byrne, and Amber Pursley, MS for helpful discussion and assistance with neuropsychological testing. This research was supported in part by grant R13-0005-04/2008 from the Polish Ministry of Science and Higher Education.




