Association studies and gene expression analyses of the DISC1-interacting molecules, pericentrin 2 (PCNT2) and DISC1-binding zinc finger protein (DBZ), with schizophrenia and with bipolar disorder†‡
Ayyappan Anitha, Kazuhiko Nakamura, and Kazuo Yamada contributed equally to this work.
How to Cite this Article: Anitha A, Nakamura K, Yamada K, Iwayama Y, Toyota T, Takei N, Iwata Y, Suzuki K, Sekine Y, Matsuzaki H, Kawai M, Thanseem I, Miyoshi K, Katayama T, Matsuzaki S, Baba K, Honda A, Hattori T, Shimizu S, Kumamoto N, Kikuchi M, Tohyama M, Yoshikawa T, Mori N. 2009. Association Studies and Gene Expression Analyses of the DISC1-Interacting Molecules, Pericentrin 2 (PCNT2) and DISC1-Binding Zinc Finger Protein (DBZ), With Schizophrenia and With Bipolar Disorder. Am J Med Genet Part B 150B:967–976.
Abstract
Disrupted-in-Schizophrenia 1 (DISC1) and its molecular cascade have been implicated in the pathophysiology of major psychoses. Previously, we identified pericentrin 2 (PCNT2) and DISC1-binding zinc finger protein (DBZ) as binding partners of DISC1; further, we observed elevated expression of PCNT2 in the postmortem brains and in the lymphocytes of bipolar disorder patients, compared to controls. Here, we examined the association of PCNT2 with schizophrenia in a case–control study of Japanese cohorts. We also examined the association of DBZ with schizophrenia and with bipolar disorder, and compared the mRNA levels of DBZ in the postmortem brains of schizophrenia, bipolar and control samples. DNA from 180 schizophrenia patients 201 controls were used for the association study of PCNT2 and DBZ with schizophrenia. Association of DBZ with bipolar disorder was examined in DNA from 238 bipolar patients and 240 age- and gender-matched controls. We observed significant allelic and genotypic associations of the PCNT2 SNPs, rs2249057, rs2268524, and rs2073380 (Ser/Arg) with schizophrenia; the association of rs2249057 (P = 0.002) withstand multiple testing correction. Several two SNP- and three SNP-haplotypes showed significant associations; the associations of haplotypes involving rs2249057 withstand multiple testing correction. No associations were observed for DBZ with schizophrenia or with bipolar disorder; further, there was no significant difference between the DBZ mRNA levels of control, schizophrenia and bipolar postmortem brains. We suggest a possible role of PCNT2 in the pathogenesis of schizophrenia. Abnormalities of PCNT2, the centrosomal protein essential for microtubule organization, may be suggested to lead to neurodevelopmental abnormalities. © 2009 Wiley-Liss, Inc.
INTRODUCTION
Several genetic studies including genome-wide linkage scans and association studies have implicated a potential susceptibility region for psychiatric disorders on chromosome 1q, especially involving the Disrupted-In-Schizophrenia 1 (DISC1) gene [Blackwood et al., 2001; Ekelund et al., 2001; Hennah et al., 2003]. DISC1 has been identified as a disrupted gene by a balanced translocation (1; 11)(q42.1; q14.3) that cosegregated with major psychiatric disorders in a large Scottish kindred [St Clair et al., 1990; Millar et al., 2000; Blackwood et al., 2001; Millar et al., 2001].
DISC1 variations have been implicated in the positive symptoms of schizophrenia [DeRosse et al., 2007; Szeszko et al., 2008], and have been reported to influence the prefrontal function [Prata et al., 2008; Szeszko et al., 2008]. DISC1-transgenic mice have been found to exhibit brain abnormalities [Kvajo et al., 2008; Pletnikov et al., 2008] and behavioral phenotypes [Clapcote et al., 2007; Hikida et al., 2007; Li et al., 2007; Pletnikov et al., 2008] reminiscent of schizophrenia. Recent studies report the association of DISC1 polymorphisms with schizophrenia [Hennah et al., 2003; Kockelkorn et al., 2004; Callicott et al., 2005; Sachs et al., 2005; Zhang et al., 2006; Chen et al., 2007; Qu et al., 2007; Saetre et al., 2008] and with bipolar disorder [Hodgkinson et al., 2004; Thomson et al., 2005].
DISC1 is a multifunctional protein capable of interacting with several cytoskeletal and centrosomal proteins via distinct functional domains [Millar et al., 2003; Miyoshi et al., 2003; Morris et al., 2003; Ozeki et al., 2003; Brandon et al., 2004]. Through these interactions, DISC1 functions as a component of the intracellular machinery that integrates multiple functions including intracellular transport, neuronal cell signaling, and neuronal migration and architecture [Hennah et al., 2006; Ishizuka et al., 2006; Porteous et al., 2006]. The impact of DISC1 across several psychiatric diagnostic categories, thus implicates a complex interaction among loci both within the gene itself, and between DISC1 and its multitude of binding partners.
Previously, we reported the fasciculation and elongation protein-zeta 1 [FEZ1; Miyoshi et al., 2003], pericentrin 2 [PCNT2; Miyoshi et al., 2004], and DISC1-binding zinc finger protein [DBZ, also known as zinc finger protein 365 (ZNF365); Hattori et al., 2007] as interacting partners of DISC1. FEZ1, PCNT2, and DBZ bind to overlapping regions of DISC1; in addition, the domain of DISC1 interacting with DBZ is close to the translocation breakpoint in DISC1 [Hattori et al., 2007]. FEZ1 has been found to be associated with schizophrenia in Japanese cohorts [Yamada et al., 2004]. Lipska et al. 2006 reported reduced mRNA levels of FEZ1 in the postmortem brain tissues of schizophrenia patients. We observed elevated expression of PCNT2 in the postmortem brains and in the peripheral blood lymphocytes (PBL) of bipolar disorder patients, compared to healthy controls; however, there was no significant association of PCNT2 with bipolar disorder [Anitha et al., 2008]. So far, there are no reports on the association of DBZ with any psychiatric disorders.
Here, we examined the genetic association of PCNT2 with schizophrenia in a case–control study of Japanese cohorts. We also examined the genetic association of DBZ with schizophrenia and with bipolar disorder. In addition, we compared the mRNA levels of DBZ in the postmortem prefrontal cortices of schizophrenia, bipolar disorder and control groups.
MATERIALS AND METHODS
Association Study
Subjects
This study was approved by the Ethics Committee of Hamamatsu University School of Medicine; patient confidentiality was maintained at all times, and written informed consent was obtained from all the participants. For the association study of PCNT2 and DBZ with schizophrenia, we collected blood samples from 180 schizophrenia patients [age 53.58 ± 12 years (mean ± SD); male/female 99:81] and 201 healthy control subjects (age 40.54 ± 13 years; male/female 98:103); there was no significant difference in sex distribution between the case and control groups (χ2 = 1.48; df = 1; P = 0.223). We examined the association of DBZ with bipolar disorder, in blood samples collected from 238 bipolar disorder patients (age 51.28 ± 13.18 years; male/female 131:107) and 240 age- and gender-matched healthy controls (age 51.49 ± 10.73 years, male/female 120:120); the controls samples were independent of the samples used in the association study of PCNT2 and DBZ with schizophrenia. All the subjects were recruited from a geographical area located in central Japan. Best-estimate lifetime diagnoses of patients were made by direct interview with experienced psychiatrists, according to DSM-IV criteria [American Psychiatric Association, 1994]. Control subjects were recruited from hospital staff and company employees documented to be free from any psychiatric problems.
Genomic DNA was extracted from whole blood using QIAamp DNA blood kit (QIAGEN, Maryland, MD).
Marker selection
The genomic structures of PCNT2 (chromosomal location 21q22.3) and DBZ (10q21.2) are based on the UCSC March 2006 draft assembly of human genome (http://www.genome.ucsc.edu). SNPs for association study were selected using the Applied Biosystems (ABI; Foster City, CA) software SNPbrowser 1.0.19; SNPs reported in databases like National Centre for Biotechnology information (NCBI dbSNP: http://www.ncbi.nlm.nih.gov/SNP) and Japanese Single Nucleotide Polymorphisms (JSNP: http://snp.ims.u-tokyo.ac.jp) were also referred to. On the basis of their genomic locations and minor allele frequencies (MAF > 0.1) in Japanese population, 19 SNPs of PCNT2 (Fig. 1) and 10 SNPs of DBZ (Fig. 2), were chosen for the association study, aiming at an average spacing of one common SNP at every 3–5 kb.
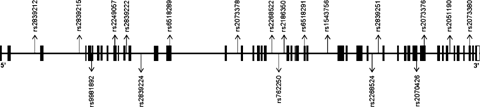
Genomic structure of PCNT2 and locations of SNPs. Exons are indicated by boxes, with translated regions in closed boxes and untranslated regions in open boxes; SNP positions are denoted by arrows.
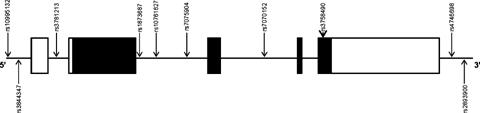
Genomic structure of DBZ and locations of SNPs. Exons are indicated by boxes, with translated regions in closed boxes and untranslated regions in open boxes; SNP positions are denoted by arrows.
Genotyping
Assay-on-Demand™ or Assay-by-Design SNP genotyping products (ABI) were used to score SNPs, based on the TaqMan assay method [Ranade et al., 2001]. Genotypes were determined in ABI PRISM 7900 Sequence Detection System (SDS), and analyzed using SDS v2.0 (ABI).
Statistical analysis
All the genotyping results were tested for Hardy–Weinberg Equilibrium (HWE). The statistical significance of variations in allelic and genotypic distributions between the schizophrenia and control groups was evaluated using Fisher's exact test. Haplotype associations were examined using the COCAPHASE program of UNPHASED v2.403 [Dudbridge, 2003; http://www.litbio.org]. Expectation maximization (EM) algorithm was used to resolve uncertain haplotypes, infer missing genotypes and provide maximum-likelihood estimation of frequencies. LD parameters were estimated using the ldmax option of GOLD v1.1.0 [Excoffier and Slatkin, 1995; Abecasis and Cookson, 2000; http://www.well.ox.ac.uk/asthma/GOLD/]. LD strength was estimated in terms of the squared correlation coefficient [r2; Devlin and Risch, 1995].
Gene Expression Analysis
Brain RNA
RNA from dorsolateral prefrontal cortex (DLPFC; Brodmann's area 46) was donated by The Stanley Medical Research Institute [SMRI; http://www.stanleyresearch.org/programs/brain_collection.asp; Torrey et al., 2000]. RNA from 31 schizophrenia patients, 33 bipolar disorder patients, and 32 control subjects were used in the study; the demographic details of each group are shown in Table I. Since the RNA samples were coded, the diagnoses of the subjects were masked, while the assays were performed.
| Variables | Control (N = 32) | Schizophrenia (N = 31) | Bipolar disorder (N = 33) | P-value |
|---|---|---|---|---|
| Age (years) (mean ± SD)a | 43.5 ± 7.39 | 42.64 ± 8.90 | 45.41 ± 10.70 | 0.457 |
| Postmortem interval (hr)a | 29.87 ± 13.30 | 31.23 ± 16.50 | 36.97 ± 17.70 | 0.164 |
| Brain pHa | 6.63 ± 0.25 | 6.47 ± 0.24 | 6.42 ± 0.30 | 0.007 |
| Male/femaleb | 23:9 | 23:8 | 16:7 | 0.039 |
| Lifetime dose of antipsychoticscd | — | 89,360 ± 105,375 | 10,339 ± 23,181 | <0.001 |
- a One-way ANOVA.
- b χ2 test.
- c Fluphenazine equivalents.
- d t-test.
- Bold values indicate significant P-values.
Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR)
Real-time qRT-PCR analysis was performed using the ABI PRISM 7900 SDS. TaqMan primer/probes for DBZ and for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which served as the endogenous reference, were purchased (Assay-on-Demand) from ABI; TaqMan assays were performed in duplicate according to the manufacturer's protocol. A comparative threshold cycle (CT) method validation experiment was done to check whether the efficiencies of target and reference amplifications were approximately equal (the slope of the log input amount vs. ΔCT < 0.1). One sample was randomly chosen as the calibrator, and was amplified in each plate, to correct for experimental differences among consecutive PCR runs. The amounts of DBZ mRNAs were normalized to the endogenous reference, and expressed relative to the calibrator as 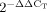 (comparative CT method).
(comparative CT method).
Statistical analysis
Statistical calculations were performed using the SPSS statistical package, version 11.0.1 (SPSS Co. Ltd., Tokyo, Japan). One-way analysis of variance (ANOVA) was used to check the variability in the distribution of demographic variables and the variability in DBZ expression across groups. Any effect of various demographic or clinical variables on DBZ expression was examined by Pearson's correlation coefficient.
RESULTS
Association Study of PCNT2 With Schizophrenia
The genotypic distributions of all the SNPs were found to be in HWE in the control group; however, in the schizophrenia group, SNP04 showed a deviation from HWE (P = 0.015). The allelic and genotypic frequencies of the 19 SNPs of PCNT2 in the schizophrenia and control groups are summarized in Table II. SNP04 (P = 0.002; OR = 1.6), SNP14 (P = 0.016; OR = 1.43), and SNP19 (P = 0.009; OR = 1.47) showed significant allelic associations with schizophrenia; the association of SNP04 withstands multiple testing correction. The frequencies of C allele of SNP04 (rs2249057; Exon 10; Ser/Ser), C allele of SNP 14 (rs2268524; Intron 31) and A allele of SNP19 (rs2073380; Exon 45; Ser/Arg) were higher in the schizophrenia group compared to controls. Further, SNP04 (P = 0.003), SNP14 (P = 0.045), and SNP19 (P = 0.025) showed significant variations in the distributions of genotypic frequencies between schizophrenia and control groups.
| Marker | SNP | Samples | Allelea | P-valueb | Genotypea | P-valueb | |||
|---|---|---|---|---|---|---|---|---|---|
| SNP01 | rs2839212 | C | T | C/C | C/T | T/T | |||
| Intron 2 | SC (180) | 261 (0.73) | 99 (0.27) | 0.111 | 96 (0.53) | 69 (0.38) | 15 (0.09) | 0.251 | |
| CT (201) | 312 (0.78) | 90 (0.22) | 121 (0.60) | 70 (0.35) | 10 (0.05) | ||||
| SNP02 | rs2839215 | G | A | G/G | G/A | A/A | |||
| Intron 3 | SC (180) | 260 (0.72) | 100 (0.28) | 0.156 | 94 (0.52) | 72 (0.40) | 14 (0.08) | 0.327 | |
| CT (201) | 309 (0.77) | 93 (0.23) | 120 (0.60) | 69 (0.34) | 12 (0.06) | ||||
| SNP03 | rs9981892 | G | A | G/G | G/A | A/A | |||
| Intron 5 | SC (178) | 254 (0.71) | 102 (0.29) | 0.135 | 90 (0.51) | 74 (0.42) | 14 (0.07) | 0.308 | |
| CT (200) | 305 (0.76) | 95 (0.24) | 116 (0.58) | 73 (0.37) | 11 (0.05) | ||||
| SNP04 | rs2249057 | C | A | C/C | C/A | A/A | |||
| Exon 10 (silent) | SC (180) | 230 (0.64) | 130 (0.36) | 0.002 | 81 (0.45) | 68 (0.38) | 31 (0.17) | 0.003 | |
| CT (201) | 211 (0.52) | 191 (0.48) | 56 (0.28) | 99 (0.49) | 46 (0.23) | ||||
| SNP05 | rs2839222 | A | G | A/A | A/G | G/G | |||
| Intron 12 | SC (178) | 258 (0.72) | 98 (0.28) | 0.075 | 93 (0.52) | 72 (0.40) | 13 (0.08) | 0.18 | |
| CT (200) | 313 (0.78) | 87 (0.22) | 122 (0.61) | 69 (0.35) | 9 (0.04) | ||||
| SNP06 | rs2839224 | G | A | G/G | G/A | A/A | |||
| Intron 13 | SC (180) | 260 (0.72) | 100 (0.28) | 0.052 | 94 (0.52) | 72 (0.40) | 14 (0.08) | 0.138 | |
| CT (200) | 314 (0.79) | 86 (0.21) | 123 (0.62) | 68 (0.34) | 9 (0.04) | ||||
| SNP07 | rs6518289 | C | T | C/C | C/T | T/T | |||
| Exon 15 (missense) | SC (180) | 258 (0.72) | 102 (0.28) | 0.114 | 94 (0.52) | 70 (0.39) | 16 (0.09) | 0.227 | |
| CT (201) | 309 (0.77) | 93 (0.23) | 118 (0.59) | 73 (0.36) | 10 (0.05) | ||||
| SNP08 | rs2073378 | C | G | C/C | C/G | G/G | |||
| Intron 16 | SC (176) | 258 (0.73) | 94 (0.27) | 0.175 | 94 (0.53) | 70 (0.40) | 12 (0.07) | 0.379 | |
| CT (201) | 312 (0.78) | 90 (0.22) | 120 (0.60) | 72 (0.36) | 9 (0.04) | ||||
| SNP09 | rs2268522 | G | A | G/G | G/A | A/A | |||
| Intron 21 | SC (180) | 245 (0.68) | 115 (0.32) | 0.204 | 83 (0.46) | 79 (0.44) | 18 (0.10) | 0.57 | |
| CT (201) | 291 (0.72) | 111 (0.28) | 103 (0.51) | 82 (0.41) | 16 (0.08) | ||||
| SNP10 | rs762250 | G | C | G/G | G/C | C/C | |||
| Intron 21 | SC (180) | 245 (0.68) | 115 (0.32) | 0.204 | 83 (0.46) | 79 (0.44) | 18 (0.10) | 0.417 | |
| CT (201) | 291 (0.72) | 111 (0.28) | 106 (0.53) | 79 (0.39) | 16 (0.08) | ||||
| SNP11 | rs2186350 | A | G | A/A | A/G | G/G | |||
| Intron 21 | SC (179) | 243 (0.68) | 115 (0.32) | 0.202 | 82 (0.46) | 79 (0.44) | 18 (0.10) | 0.392 | |
| CT (199) | 288 (0.72) | 110 (0.28) | 105 (0.53) | 78 (0.39) | 16 (0.08) | ||||
| SNP12 | rs6518291 | A | G | A/A | A/G | G/G | |||
| Exon 26 (missense) | SC (180) | 262 (0.73) | 98 (0.27) | 0.051 | 95 (0.53) | 72 (0.40) | 13 (0.07) | 0.149 | |
| CT (201) | 317 (0.79) | 85 (0.21) | 125 (0.62) | 67 (0.33) | 9 (0.05) | ||||
| SNP13 | rs1543756 | G | A | G/G | G/A | A/A | |||
| Intron 27 | SC (180) | 262 (0.73) | 98 (0.27) | 0.051 | 95 (0.53) | 72 (0.40) | 13 (0.07) | 0.149 | |
| CT (201) | 317 (0.79) | 85 (0.21) | 125 (0.62) | 67 (0.33) | 9 (0.05) | ||||
| SNP14 | rs2268524 | C | T | C/C | C/T | T/T | |||
| Intron 31 | SC (178) | 211 (0.59) | 145 (0.41) | 0.016 | 63 (0.35) | 85 (0.48) | 30 (0.17) | 0.045 | |
| CT (201) | 203 (0.50) | 199 (0.50) | 49 (0.24) | 105 (0.52) | 47 (0.24) | ||||
| SNP15 | rs2839251 | C | T | C/C | C/T | T/T | |||
| Intron 31 | SC (179) | 260 (0.73) | 98 (0.27) | 0.211 | 94 (0.53) | 72 (0.40) | 13 (0.07) | 0.434 | |
| CT (201) | 308 (0.77) | 94 (0.23) | 117 (0.58) | 74 (0.37) | 10 (0.05) | ||||
| SNP16 | rs2070426 | C | G | C/C | C/G | G/G | |||
| Exon 37 (missense) | SC (179) | 243 (0.68) | 115 (0.32) | 0.176 | 82 (0.46) | 79 (0.44) | 18 (0.10) | 0.364 | |
| CT (200) | 290 (0.73) | 110 (0.27) | 105 (0.53) | 80 (0.40) | 15 (0.07) | ||||
| SNP17 | rs2073376 | G | A | G/G | G/A | A/A | |||
| Exon 38 (missense) | SC (179) | 243 (0.68) | 115 (0.32) | 0.176 | 82 (0.46) | 79 (0.44) | 18 (0.10) | 0.364 | |
| CT (200) | 290 (0.73) | 110 (0.27) | 105 (0.53) | 80 (0.40) | 15 (0.07) | ||||
| SNP18 | rs2051190 | T | C | T/T | T/C | C/C | |||
| Intron 41 | SC (180) | 245 (0.68) | 115 (0.32) | 0.177 | 83 (0.46) | 79 (0.44) | 18 (0.10) | 0.374 | |
| CT (201) | 292 (0.73) | 110 (0.27) | 106 (0.53) | 80 (0.40) | 15 (0.07) | ||||
| SNP19 | rs2073380 | A | C | A/A | A/C | C/C | |||
| Exon 45 (missense) | SC (180) | 216 (0.60) | 144 (0.40) | 0.009 | 66 (0.37) | 84 (0.47) | 30 (0.16) | 0.025 | |
| CT (200) | 202 (0.50) | 198 (0.50) | 49 (0.25) | 104 (0.52) | 47 (0.23) | ||||
- SC, schizophrenia; CT, control; the number of genotyped individuals are given in parentheses.
- a Number followed by frequency in parentheses.
- b Fisher's Exact test; significant P-values are indicated in bold italics.
The P-values of haplotypic distributions between schizophrenia and control groups, involving groups of two SNPs (with one SNP overlap) and groups of three SNPs (with two SNP overlap) are given in Table III. Several two-SNP haplotypes showed significant associations; among these, the association of SNP03–SNP04 (P = 0.001) reached Bonferroni-corrected significance [0.0027 (α/number of comparisons = 0.05:18)]. Several three-SNP haplotypes also showed significant associations, among which, the association of SNP03–SNP04–SNP05 (P = 0.002) reached Bonferroni-corrected significance [0.0029 (α/number of comparisons = 0.05:17)].
| Two-SNP | P-valuea | Three-SNP | P-valuea |
|---|---|---|---|
| 1-2 | 0.182 | 1-2-3 | 0.057 |
| 2-3 | 0.049 | 2-3-4 | 0.019 |
| 3-4 | 0.001 | 3-4-5 | 0.002 |
| 4-5 | 0.006 | 4-5-6 | 0.004 |
| 5-6 | 0.043 | 5-6-7 | 0.086 |
| 6-7 | 0.091 | 6-7-8 | 0.297 |
| 7-8 | 0.199 | 7-8-9 | 0.129 |
| 8-9 | 0.066 | 8-9-10 | 0.044 |
| 9-10 | 0.241 | 9-10-11 | 0.225 |
| 10-11 | 0.178 | 10-11-12 | 0.008 |
| 11-12 | 0.008 | 11-12-13 | 0.055 |
| 12-13 | 0.055 | 12-13-14 | 0.047 |
| 13-14 | 0.039 | 13-14-15 | 0.004 |
| 14-15 | 0.061 | 14-15-16 | 0.246 |
| 15-16 | 0.038 | 15-16-17 | 0.03 |
| 16-17 | 0.162 | 16-17-18 | 0.162 |
| 17-18 | 0.164 | 17-18-19 | 0.017 |
| 18-19 | 0.023 |
- a Significant P-values are indicated in bold italics.
A graphic representation of the LD strength (r2 values) between markers in the schizophrenia group is shown in Figure S1; strong LD (r2 = 0.920) was observed between SNPs 14 and 19, which showed associations with schizophrenia. The LD pattern was found to be similar in the schizophrenia and control (data not shown) groups.
Association Study of DBZ With Schizophrenia
The allelic and genotypic frequencies of the 10 SNPs of DBZ are given in Table IV. Genotypic distributions of all the SNPs were found to be in HWE, in the schizophrenia and control groups. None of the SNPs showed any significant associations with schizophrenia. No significant haplotype associations were observed in the two-SNP or three-SNP haplotype analyses.
| Marker | SNP | Samples | Allelea | P-valueb | Genotypea | P-valueb | |||
|---|---|---|---|---|---|---|---|---|---|
| SNP01 | rs10995132 | G | A | G/G | G/A | A/A | |||
| 5′ | SC (179) | 311 (0.87) | 47 (0.13) | 0.660 | 135 (0.75) | 41 (0.23) | 3 (0.02) | 0.794 | |
| CT (199) | 345 (0.86) | 47 (0.14) | 148 (0.74) | 49 (0.24) | 2 (0.02) | ||||
| SNP02 | rs3844347 | A | G | A/A | A/G | G/G | |||
| 5′ | SC (180) | 288 (0.80) | 72 (0.20) | 0.426 | 116 (0.65) | 56 (0.31) | 8 (0.04) | 0.541 | |
| CT (201) | 311 (0.77) | 91 (0.23) | 119 (0.59) | 73 (0.36) | 9 (0.05) | ||||
| SNP03 | rs3781213 | G | C | G/G | G/C | C/C | |||
| Intron 1 | SC (180) | 287 (0.79) | 73 (0.21) | 0.428 | 115 (0.64) | 57 (0.31) | 8 (0.05) | 0.561 | |
| CT (201) | 310 (0.77) | 92 (0.23) | 118 (0.59) | 74 (0.36) | 9 (0.05) | ||||
| SNP04 | rs1873687 | T | C | T/T | T/C | C/C | |||
| Intron 2 | SC (180) | 285 (0.79) | 75 (0.21) | 0.659 | 114 (0.63) | 57 (0.32) | 9 (0.05) | 0.560 | |
| CT (200) | 311 (0.78) | 89 (0.22) | 119 (0.60) | 73 (0.36) | 8 (0.04) | ||||
| SNP05 | rs10761627 | T | C | T/T | T/C | C/C | |||
| Intron 2 | SC (180) | 285 (0.79) | 75 (0.21) | 0.539 | 114 (0.63) | 57 (0.32) | 9 (0.05) | 0.595 | |
| CT (200) | 309 (0.77) | 91 (0.23) | 118 (0.59) | 73 (0.37) | 9 (0.04) | ||||
| SNP06 | rs7075904 | C | G | C/C | C/G | G/G | |||
| Intron 2 | SC (180) | 309 (0.86) | 51 (0.14) | 0.917 | 133 (0.74) | 43 (0.24) | 4 (0.02) | 1.000 | |
| CT (200) | 345 (0.86) | 55 (0.14) | 149 (0.74) | 47 (0.24) | 4 (0.02) | ||||
| SNP07 | rs7070152 | A | G | A/A | A/G | G/G | |||
| Intron 3 | SC (180) | 286 (0.79) | 74 (0.21) | 0.857 | 116 (0.64) | 54 (0.30) | 10 (0.06) | 0.544 | |
| CT (201) | 322 (0.80) | 80 (0.20) | 128 (0.64) | 66 (0.33) | 7 (0.03) | ||||
| SNP08 | rs3758490 | T | G | T/T | T/G | G/G | |||
| Exon 5 (Ser/Ala) | SC (176) | 221 (0.63) | 131 (0.37) | 0.063 | 66 (0.37) | 89 (0.51) | 21 (0.12) | 0.143 | |
| CT (199) | 276 (0.69) | 122 (0.31) | 93 (0.47) | 90 (0.45) | 16 (0.08) | ||||
| SNP09 | rs4746698 | A | G | A/A | A/G | G/G | |||
| 3′ | SC (179) | 246 (0.69) | 112 (0.31) | 0.641 | 87 (0.49) | 72 (0.40) | 20 (0.11) | 0.301 | |
| CT (200) | 268 (0.67) | 132 (0.33) | 86 (0.43) | 96 (0.48) | 18 (0.09) | ||||
| SNP10 | rs2893900 | C | T | C/C | C/T | T/T | |||
| 3′ | SC (179) | 337 (0.94) | 21 (0.06) | 0.207 | 159 (0.89) | 19 (0.10) | 1 (0.01) | 0.249 | |
| CT (201) | 368 (0.91) | 34 (0.09) | 168 (0.83) | 32 (0.16) | 1 (0.01) | ||||
- SC, schizophrenia; CT, control; the number of genotyped individuals are given in parentheses.
- a Number followed by frequency in parentheses.
- b Fisher's Exact test.
Association Study of DBZ With Bipolar Disorder
The allelic and genotypic frequencies of the 10 SNPs of DBZ are shown in Table V. The genotypic distributions of all the SNPs were found to be in HWE in the control group; however, SNP09 (rs4746698) showed a marginal deviation from HWE, in the bipolar group (P = 0.033). None of the SNPs showed any significant associations with bipolar disorder. No significant haplotype associations were observed in the two-SNP or three-SNP haplotype analyses.
| Marker | SNP | Samples | Allelea | P-valueb | Genotypea | P-valueb | |||
|---|---|---|---|---|---|---|---|---|---|
| SNP01 | rs10995132 | G | A | G/G | G/A | A/A | |||
| 5′ | BD (237) | 407 (0.86) | 67 (0.14) | 0.783 | 174 (0.73) | 59 (0.25) | 4 (0.02) | 0.801 | |
| CT (240) | 409 (0.85) | 71 (0.15) | 172 (0.72) | 65 (0.27) | 3 (0.01) | ||||
| SNP02 | rs3844347 | A | G | A/A | A/G | G/G | |||
| 5′ | BD (238) | 372 (0.78) | 104 (0.22) | 0.489 | 146 (0.61) | 80 (0.34) | 12 (0.05) | 0.565 | |
| CT (240) | 366 (0.76) | 114 (0.24) | 137 (0.57) | 92 (0.38) | 11 (0.05) | ||||
| SNP03 | rs3781213 | G | C | G/G | G/C | C/C | |||
| Intron 1 | BD (238) | 367 (0.77) | 109 (0.23) | 0.818 | 143 (0.60) | 81 (0.34) | 14 (0.06) | 0.443 | |
| CT (239) | 365 (0.76) | 113 (0.24) | 136 (0.57) | 93 (0.39) | 10 (0.04) | ||||
| SNP04 | rs1873687 | T | C | T/T | T/C | C/C | |||
| Intron 2 | BD (236) | 368 (0.78) | 104 (0.22) | 0.489 | 145 (0.62) | 78 (0.33) | 13 (0.05) | 0.419 | |
| CT (240) | 365 (0.76) | 115 (0.24) | 136 (56.67) | 93 (38.75) | 11 (4.58) | ||||
| SNP05 | rs10761627 | T | C | T/T | T/C | C/C | |||
| Intron 2 | BD (237) | 368 (0.78) | 106 (0.22) | 0.591 | 144 (0.61) | 80 (0.34) | 13 (0.05) | 0.540 | |
| CT (239) | 364 (0.76) | 114 (0.24) | 136 (0.57) | 92 (0.38) | 11 (0.05) | ||||
| SNP06 | rs7075904 | C | G | C/C | C/G | G/G | |||
| Intron 2 | BD (237) | 405 (0.85) | 69 (0.15) | 0.927 | 173 (0.73) | 59 (0.25) | 5 (0.02) | 0.676 | |
| CT (240) | 409 (0.85) | 71 (0.15) | 172 (0.72) | 65 (0.27) | 3 (0.01) | ||||
| SNP07 | rs7070152 | A | G | A/A | A/G | G/G | |||
| Intron 3 | BD (237) | 385 (0.81) | 89 (0.19) | 0.333 | 159 (0.67) | 67 (0.28) | 11 (0.05) | 0.156 | |
| CT (240) | 377 (0.78) | 103 (0.22) | 145 (0.61) | 87 (0.36) | 8 (0.03) | ||||
| SNP08 | rs3758490 | T | G | T/T | T/G | G/G | |||
| Exon 5 (Ser/Ala) | BD (236) | 323 (0.68) | 149 (0.32) | 0.445 | 115 (0.49) | 93 (0.39) | 28 (0.12) | 0.242 | |
| CT (240) | 317 (0.66) | 163 (0.34) | 102 (0.43) | 113 (0.47) | 25 (0.10) | ||||
| SNP09 | rs4746698 | A | G | A/A | A/G | G/G | |||
| 3′ | BD (235) | 323 (0.69) | 147 (0.31) | 0.333 | 118 (0.50) | 87 (0.37) | 30 (0.13) | 0.086 | |
| CT (240) | 315 (0.66) | 165 (0.34) | 101 (0.42) | 113 (0.47) | 26 (0.11) | ||||
| SNP10 | rs2893900 | C | T | C/C | C/T | T/T | |||
| 3′ | BD (238) | 438 (0.92) | 38 (0.08) | 0.364 | 202 (0.85) | 34 (0.14) | 2 (0.01) | 0.633 | |
| CT (240) | 433 (0.90) | 47 (0.10) | 196 (0.82) | 41 (0.17) | 3 (0.01) | ||||
- BD, bipolar disorder; CT, control; the number of genotyped individuals are given in parentheses.
- a Number followed by frequency in parentheses.
- b Fisher's Exact test.
DBZ Gene Expression Analysis
The variability in the distribution of demographic variables across the control, schizophrenia and bipolar disorder groups are summarized in Table I. There was no significant difference (F = 0.34; df = 2.93; P = 0.713) in DBZ expression across the three groups (Fig. S2). No significant correlations were observed between DBZ expression and any of the clinical features of schizophrenia or bipolar groups.
DISCUSSION
In this study, we observed SNP- and haplotype- associations of the DISC1-interacting molecule PCNT2 with schizophrenia. SNP04, SNP14, and SNP19 showed significant allelic and genotypic associations. The allelic association of SNP04 and the haplotypic associations involving SNP04 withstand multiple testing correction. Deviation from HWE was observed for SNP04 in schizophrenia patients; heterozygotes of patients were lower than the expected values based on HWE. Since SNP04 showed association, which withstands correction for multiple testing, the deviation from HWE in patients, but not in controls, may be viewed as an additional evidence of association. We observed a strong LD between SNPs 14 and 19. SNP19, located in exon 45, is a non-synonymous SNP with Ser/Arg substitution. Since Ser is a neutral amino acid and Arg is a strong basic amino acid, the substitution may be suggested to exert an influence on the protein structure of PCNT2. To the best of our knowledge, this is the first report of a genetic association study of PCNT2 with schizophrenia.
Abnormalities of brain morphogenesis, especially cortical dysplasia, have been observed in individuals with partial deletion of chromosome 21q22.3 harboring the PCNT2 gene [Yao et al., 2006]. PCNT2, an integral component of the pericentriolar material, interacts with pericentriolar material 1 (PCM1), another member of the DISC1 interactome [Kamiya et al., 2008], for its centrosomal localization, and to accomplish its function in microtubule organization [Balczon et al., 1994; Doxsey et al., 1994; Kubo et al., 1999; Dammermann and Merdes, 2002]. Recently, PCM1 has been implicated in susceptibility to schizophrenia [Gurling et al., 2006]. In the developing cerebral cortex, suppression of PCM1 has been reported to lead to neuronal migration defects [Kamiya et al., 2008]. Thus, centrosomal proteins such as PCNT2 may be suggested to have an important role in cortical development, and therefore, in the pathogenesis of neurodevelopmental disorders like schizophrenia. The PCNT2-binding region of DISC1 overlaps with the region interacting with FEZ1 [Miyoshi et al., 2004], a schizophrenia susceptibility gene that plays a vital role in axonal outgrowth and fasciculation [Yamada et al., 2004].
In our previous study, elevated expression of PCNT2 was observed in the brain and in the PBL of bipolar disorder patients, compared to controls; however, there was no significant difference in PCNT2 expression between control and schizophrenia groups [Anitha et al., 2008]. We suggested that a possible apoptotic process in the brain, resulting from over-expression of PCNT2, may underlie the pathogenesis of bipolar disorder. However, in the subsequent association study, none of the SNPs showed association with bipolar disorder.
Considering the results from the genetic analyses of PCNT2 in bipolar disorder and schizophrenia, it might be suggested that DISC1 and its interacting molecules are involved in psychiatric symptoms that cross diagnostic boundaries. The translocation in the original Scottish family [St Clair et al., 1990] demonstrated that the disruption of DISC1 gene, although sufficient to predispose an individual to psychiatric disorder, was, in itself, insufficient to predict any particular disorder. Although the family members showed predominantly schizophrenic symptoms, they also manifested a wide spectrum of other psychiatric phenotypes. In addition to schizophrenia, DISC1 has also been found to be associated with other neuropsychiatric disorders including bipolar disorder [Hodgkinson et al., 2004; Thomson et al., 2005], major depression [Hashimoto et al., 2006], and autism [Kilpinen et al., 2008]. Accumulating evidence show that malfunction of DISC1 lead to brain structural and functional abnormalities [Faulkner et al., 2008; Kvajo et al., 2008; Pletnikov et al., 2008; Prata et al., 2008; Szeszko et al., 2008]. Thus, it may be suggested that the basis of the shared risk for major neurodevelopmental disorders could be centered on DISC1, while its interaction with other members of the interactome lead to different phenotypic outcomes.
Several other DISC1-interacting molecules, including phosphodiesterase 4B [PDE4B; Numata et al., 2008], 14-3-3ε [YWHAE; Ikeda et al., 2008], nudE nuclear distribution gene E homolog 1 [NDE1; Hennah et al., 2007], nudE nuclear distribution gene E homolog like 1 [NDEL1; Burdick et al., 2008], and activating transcription factor 4 [ATF4; Qu et al., 2008], have been suggested as potential susceptibility genes for schizophrenia. The DISC1 interactome is expanding with the discovery of novel interacting proteins, and this information could be useful in the identification of potential pathways involving DISC1, in the pathology of major psychoses.
In this study, no significant genetic associations were observed for DBZ, the other DISC1-interacting molecule, with schizophrenia or with bipolar disorder; in addition, there was no significant difference between the DBZ mRNA levels of control, schizophrenia and bipolar postmortem brains.
In conclusion, we suggest a possible role of PCNT2 in the pathogenesis of schizophrenia. Abnormalities of PCNT2 may lead to defects in microtubule function; during the development of the central nervous system, this dysfunction might result in alterations in neuronal migration, axonal extension, and neurite outgrowth, subsequently leading to impaired neurodevelopment. Our study is limited by small sample size; therefore, the results should be treated with caution, until replication using a larger sample size. It would also be interesting to study the role of PCNT2 in neurodevelopment.
Acknowledgements
Postmortem brain tissues were donated by the Stanley Medical Research Institute's brain collection, courtesy of Dr. Michael B Knable, Dr. E Fuller Torrey, Dr. Maree J Webster, Dr. Serge Weis, and Dr. Robert H Yolke. This work was supported by a Grant-in-Aid for Scientific Research (B) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. We thank Hosoya Teruyo for technical assistance.




