G72/G30 (DAOA) and juvenile-onset mood disorders†
How to Cite this Article: Gomez L, Wigg K, Feng Y, Kiss E, Kapornai K, Tamás Z, Mayer L, Baji I, Daróczi G, Benák I, Kothencné VO, Dombovári E, Kaczvinszk E, Besnyő M, Gádoros J, King N, Székely J, Kovacs M, Vetró Á, Kennedy JL, Barr CL. 2008. G72/G30 (DAOA) and juvenile-onset mood disorders. Am J Med Genet Part B 150B:1007–1012.
Abstract
The chromosome 13q region has been linked to bipolar disorder in a number of genome scans as well as focused linkage studies. Previously we identified linkage to the 13q32 region in a genome scan of 146 affected sibling pair families from Hungary with juvenile-onset mood disorders. Within this region are the overlapping genes G72/G30, with G72 now officially named as D-amino-acid oxidase activator (DAOA). This locus has been associated with panic disorder, schizophrenia, and bipolar disorder. In this study, we tested for association to 11 markers in these genes and mood disorders in a sample of 646 nuclear families identified with a proband with onset of a mood disorder before 14.9 years of age. We identified evidence for association to three markers within the gene (rs2391191, rs3918341, rs1935062), two of which had been associated with bipolar disorder in previous studies. When corrected for the number of markers tested, the results were no longer significant, however the prior evidence for association of this gene in multiple studies points to this gene as a potential contributor to juvenile-onset mood disorders. © 2008 Wiley-Liss, Inc.
Mood disorders (bipolar and depressive disorders) in children and adolescents are prevalent and associated with high morbidity and mortality [Kessler et al., 2003]. Previously, we performed a genome scan on 146 nuclear families from Hungary with affected sibling pairs diagnosed with juvenile-onset mood disorders [Wigg et al., submitted]. Although no marker provided evidence for a novel linkage finding at the recommended genome wide significant level, we identified a region with nominal evidence for linkage (P < 0.01) on chromosome 13q. This region has been linked to mood disorders (bipolar disorder and recurrent depressive disorder) in previous studies [Stine et al., 1997; Detera-Wadleigh et al., 1999; Kelsoe et al., 2001; Liu et al., 2001; Shaw et al., 2003], thus the results met the criteria for significance as a replicated linkage region [Lander and Kruglyak, 1995]. The distal 13q region has been also linked to other psychiatric disorders and phenotypes including schizophrenia (13q32) [Blouin et al., 1998; Brzustowicz et al., 1999], panic disorder [Hamilton et al., 2003], mood-incongruent psychotic features in bipolar disorder (13q21-33) [Goes et al., 2007], and psychotic bipolar disorder (13q32, 13q31) [Park et al., 2004].
Localized in the 13q linkage region are the genes G72, also termed d-serine amino acid oxidase activator (DAOA), and G30 which are overlapping genes that are transcribed in opposite directions and span ∼29 and ∼47 kb of genomic sequence, respectively. The G72/G30 locus was first identified as associated with schizophrenia [Chumakov et al., 2002], a finding that has been replicated [Abou Jamra et al., 2006; Craddock et al., 2006; Detera-Wadleigh and McMahon 2006; Shi et al., 2008]. However, the association findings are far from unanimous and further are also not consistent across the associated markers or alleles [Abou Jamra et al., 2006; Craddock et al. 2006; Detera-Wadleigh and McMahon 2006; Shi et al. 2008]. Following the reports of association of G72/G30 with schizophrenia, five studies [Hattori et al., 2003; Chen et al., 2004; Schumacher et al., 2004; Fallin et al., 2005; Williams et al., 2006] and a meta-analysis [Detera-Wadleigh and McMahon, 2006] have found association of G72/G30 with bipolar disorder.
The function of G72/G30 is unknown. Until now, only two studies have investigated the function of G72 [Chumakov et al., 2002; Kvajo et al., 2007]. The G30 transcript appears not to be translated [Chumakov et al., 2002], suggesting a different function for G30, although a role in regulation of G72 is a possibility. The first study of G72 demonstrated that the G72 protein binds to, and activates, d-amino acid oxidase (DAO) in vitro [Chumakov et al., 2002], an enzyme that oxidizes d-serine, an agonist of N-methyl-d-aspartate type glutamate (NMDA) receptor [Chumakov et al., 2002].
A very recent study of the function of G72, however, failed to confirm the proposed physical interaction between G72 and DAOA [Kvajo et al., 2007]. Instead, they showed that one splicing isoform of the gene encodes a mitochondrial protein, with further evidence for a role in modulating the morphology of the mitochondrial network affecting mitochondrial operations such as oxidative phosporylation, regulation of apoptosis, and mitochondrial transport. That study also reported over expression of G72 in immature primary neurons is accompanied by a robust increase in dendritic arborization [Kvajo et al., 2007].
In this article, we follow up our linkage findings in the genome scan previously performed in sibling pairs with juvenile-onset mood disorders by testing for association with 11 polymorphisms in the gene for G72/G30 using a large sample of 646 Hungarian nuclear families, recruited from 23 mental health facilities across Hungary, with a proband with onset of a mood disorder before 15 years of age. This sample consisted of the sample of 146 sibpairs linked to this region used in the previous genome scan as well as an additional 500 parent/child trios with a childhood-onset proband. The diagnostic assessment, and inclusion/exclusion criteria for these families has been previously reported in detail [Liu et al., 2006; Kapornai et al., 2007; Kiss et al., 2007]. All children were between the ages of 7 and 14.9 years of age when they were first evaluated for this study. Twenty of the siblings genotyped for this study were unaffected at 14.9 years of age but experienced a depressive episode before the age of 18. The majority of the subjects (87%) were in a current episode of depression at their first assessment. Written informed consent for adults and assent for children was obtained from all participants.
A total of 12 SNPs were genotyped in the entire sample, with 11 of these SNPs across G72/G30. We also genotyped a marker in the VGCNL1 gene because of the prior evidence for association this marker provided in a whole genome association study [Baum et al., 2007]. These polymorphisms were genotyped by standard techniques using TaqMan® 5′ nuclease assays with primers and probes available commercially (Applied Biosystems, Foster City, CA, Assay-on-Demand) or designed specifically for this study (Applied Biosystems, Assay-by-Design). Genotyping errors were checked by first identifying Mendelian errors using the PedStats program (http://www.sph.umich.edu/csg/abecasis/PedStats/). Further data checking was performed using the error option of Merlin (http://www.sph.umich.edu/csg/abecasis/Merlin/) [Abecasis et al., 2002] to identify potential double recombinants as a sensitive check for genotyping errors. All Mendelian errors and double recombinants were either resolved or removed from the analyses. The TDTphase program from the UNPHASED package [Dudbridge, 2003] was used to test for the biased transmission of alleles for single markers and the TRANSMIT program [Clayton, 1999] for the transmission of haplotypes. The robust estimator options were used for the analyses because of the prior evidence for linkage to this region and the use of siblings in the analyses. Haplotypes with frequencies less than 0.10 were pooled for the analyses. The degree of linkage disequilibrium (LD) between marker alleles in this study was evaluated using Haploview v3.11 (http://www.broad.mit.edu/mpg/haploview) [Barrett et al., 2005].
Some of the markers genotyped across the G72/G30 locus (rs1935058, rs1341402, rs2391191, rs3918341, rs1935062, rs947267, and rs3918342) have been previously analyzed in association studies of this gene with schizophrenia and/or bipolar disorder [Detera-Wadleigh and McMahon, 2006]. The majority of these showed significant evidence for association to bipolar disorder in independent studies (rs1935058, rs1341402, rs2391191, rs1935062, and rs947267) and/or the meta-analysis (rs1935058, rs1935062, and rs3918342) [Detera-Wadleigh and McMahon, 2006]. Looking for a replication of previous findings, these markers were selected for this study. We were unable to design a genotyping assay for rs1935058, despite several different primer/probe designs. The remainder of the markers (rs12874006, rs1539070, rs9284226, rs2153674, and rs9558567) were selected using the Tagger software (version 1.2.5, http://www.broad.mit.edu/mpg/tagger) [de Bakker et al., 2005] as complementary tag SNPs for covering the variation in the entire gene, after forcing the previously studied markers to be included in the selection. For this function, Tagger used the International HapMap Project's LD data from individuals of the CEU sample (http://www.hapmap.org; Release 21/phase II Jul06, on NCBI B35 assembly, dbSNP b125). No new markers in this gene have been added in the more recently release of this project. One marker (rs9513877) within the voltage-gated ion channel like 1 (VCGNL1) gene located ∼4 Mb from G72/G30 was also genotyped in the same sample based on an association report from a whole genome association study of bipolar disorder [Baum et al., 2007]. All markers were in Hardy–Weinberg equilibrium.
Allele frequencies for these markers in the parental chromosomes were similar to those reported in European Caucasian samples with the exception of the marker rs9284226. We observed the minor allele frequency (MAF) to be 0.346 whereas the MAF for this marker reported in the HapMap Project CEU sample was 0.064. No Mendelian errors were detected and no crossovers were observed between this marker and other markers in the gene. Further, LD between this marker and the other markers was very high indicating that this marker is located in G72/G30. We further sequenced the PCR product from select individuals representing all three genotypes and verified that the correct polymorphism was amplified. Thus we confirmed the identity of the polymorphisms and we have no explanation as to why the alleles differ so dramatically from the reported alleles for Caucasians in HapMap. In any case, there was no evidence for biased transmission of the alleles of this marker to affected children.
Using TDT analysis, we did not find significant evidence for biased transmission of alleles at the marker rs9513877 in VGCNL1 (χ2 = 1.519, 2 d.f., P = 0.218). However, for the TDT analyses of the markers in G72/G30, we observed significant evidence for association between mood disorders and three markers, rs2391191, rs3918341, and rs1935062 (Table I). We used a conservative correction (robust estimator) for these analyses to account for the inclusion of the linkage families in the association studies. Permutation testing to correct for the number of markers reduced the significance such that the results for the most significant single marker were no longer significant (P = 0.18). We also analyzed the sample of families that were used in the genome scan separated from the remainder of the families and the results for both samples were less significant than the combined sample with trends in both samples, indicating the association signal is coming from both the affected sibling pair families and the trios.
| Gene | Locationa | Marker | Chromosome positiona | Allele | Allele frequency | Transmissionsb | Non-transmissions | χ2 | P-value |
|---|---|---|---|---|---|---|---|---|---|
| G72/G30 | 5′ | rs1341402 | 104913510 | Ccd | 0.224 | 192 | 183 | 0.216 | 0.642 |
| Te | 0.776 | 183 | 192 | ||||||
| Exon 2 | rs2391191 | 104917447 | A Lys | 0.373 | 232 | 283 | 5.059 | 0.025 | |
| G Argd | 0.627 | 283 | 232 | ||||||
| Intron 2 | rs3918341 | 104918420 | A | 0.571 | 298 | 247 | 4.779 | 0.029 | |
| G | 0.429 | 247 | 298 | ||||||
| Intron 2 | rs12874006 | 104918986 | G | 0.889 | 99 | 112 | 0.801 | 0.371 | |
| T | 0.111 | 112 | 99 | ||||||
| Intron 2 | rs1539070 | 104922458 | C | 0.828 | 137 | 146 | 0.286 | 0.593 | |
| G | 0.172 | 146 | 137 | ||||||
| Intron 3 | rs1935062 | 104926137 | Tf | 0.643 | 267 | 218 | 4.959 | 0.026 | |
| Ge | 0.357 | 218 | 267 | ||||||
| Intron 3 | rs9284226 | 104928138 | A | 0.346 | 247 | 221 | 1.445 | 0.229 | |
| G | 0.654 | 221 | 247 | ||||||
| Intron 3 | rs2153674 | 104929139 | C | 0.442 | 271 | 268 | 0.017 | 0.897 | |
| T | 0.558 | 268 | 271 | ||||||
| Intron 3 | rs9558567 | 104929595 | T | 0.646 | 266 | 225 | 3.428 | 0.064 | |
| A | 0.354 | 225 | 266 | ||||||
| Intron 3 | rs947267 | 104937663 | Gf | 0.423 | 270 | 248 | 0.934 | 0.334 | |
| T | 0.577 | 248 | 270 | ||||||
| 3′ | rs3918342 | 104983750 | Gg | 0.492 | 278 | 253 | 1.177 | 0.278 | |
| A | 0.508 | 253 | 278 | ||||||
| VGCNL1 | Intron | rs9513877 | 100724340 | A | 0.368 | 244 | 272 | 1.519 | 0.218 |
| G | 0.632 | 272 | 244 |
- Significant P-values are given in bold numerals. P-values shown are not corrected for multiple testing.
- a Based on NCBI Build 36.1, the March 2006 UCSC human reference sequence.
- b Number of times the alleles were transmitted from heterozygous parents to affected offspring in this sample.
- c Associated with bipolar disorder in Williams et al. 2006.
- d Associated with major mood disorder in Williams et al. 2006.
- e Associated with bipolar disorder in Hattori et al. 2003.
- f Associated with bipolar disorder in Chen et al. 2004.
- g Associated with bipolar disorder in Schumacher et al. 2004.
Very high LD was observed across markers in the G72/G30 genes as previously reported [Korostishevsky et al., 2004; Williams et al., 2006] and as observed in HapMap (http://www.hapmap.org/). In our data, high LD is seen between the first 10 proximal markers in the gene (rs1341402 through rs947267) with low LD observed between these markers and the most distal marker genotyped (rs3918342) located outside of the gene region (Fig. 1). Haplotype analyses of the 10 markers in strong LD did not identify a stronger association finding than single marker analyses as previously reported [Williams et al., 2006].
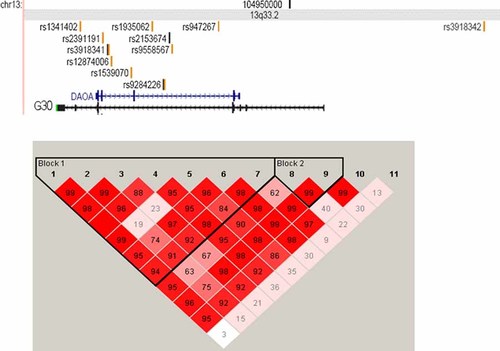
Position of genotyped markers and inter-marker LD. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
We also examined the LD between the marker in VGCNL1 and markers in G72/G30 to see if there is any relationship that could explain the previous evidence for association to this marker in the whole genome association. However, in our sample, there was no evidence for LD between the VGCNL1 marker and markers in G72/G30 for any marker combination.
While there is a significant evidence for association to schizophrenia, bipolar disorder, as well as modest association to panic disorder [Schumacher et al., 2005] to this locus, the association findings differ across studies for the associated markers as well as the associated alleles [Abou Jamra et al., 2006; Detera-Wadleigh and McMahon, 2006]. Further, haplotype analyses indicate association to different haplotypes, or no evidence for association to haplotypes [Abou Jamra et al., 2006]. These combined results suggest the possibility of allelic heterogeneity across samples or across diagnostic criteria as currently defined. Alternatively, G72 may not be the risk gene and the lack of strong consistent association signal may indicate a neighboring gene with LD to these markers as the risk gene. Although we tested the single marker in VGCNL1 with evidence for association from a genome scan, we cannot rule out this gene as a susceptible locus.
Meta-analysis of the three studies of this locus and bipolar disorder up to January 2006, indicated three associated markers across studies; rs1935058 (combined P = 0.0153), rs1935062 (combined P = 0.0019), rs3918342 (combined P = 0.0309) [Detera-Wadleigh and McMahon, 2006]. However, this meta-analysis did not distinguish the association with the different alleles at these markers. The study of Williams et al. 2006 published after that meta-analysis manuscript, identified evidence for association with three markers (rs3916956 (M12) G allele P = 0.047; rs1341402 C allele, P = 0.03; DAOA_3′UTR_SNP12 A allele P = 0.01) in a sample of cases with bipolar disorder but not a sample with schizophrenia. Further, they identified association with four markers (rs3916956 (M12) G allele P = 0.02; rs1341402 C allele P = 0.01; rs2391191 (M15) C allele P = 0.02; DAOA_3′UTR_SNP12 A allele P = 0.002) in the subset of cases with episodes of major depression but not to cases with psychotic features. They conclude that this locus influences susceptibility to mood disorders across these diagnoses. The most recent meta-analysis, that analyzed the association of specific alleles, found no evidence for association to the alleles and bipolar disorder for any of the five single markers with significant evidence for heterogeneity between studies in European samples as well as study design [Shi et al., 2008].
By comparison to the published association findings in bipolar disorder we did not find evidence for association for the markers rs3918342 or rs1341402. We did however find evidence for biased transmission of the T(A) allele of rs1935062 in agreement with the study of Chen et al. 2004 and for biased transmission of the G(C) allele for rs2391191 (M15) in agreement with the association findings of Williams et al. 2006. While our association results were quite modest, and did not stand up to correction for multiple tests for the markers used, the prior evidence for this gene as a susceptibility gene in mood disorders supports a role for this gene in juvenile-onset mood disorders. At the time of their assessment, less than 1% of the children met the criteria for bipolar disorder. However, previous longitudinal studies of similar samples, indicates that approximately 15–30% of these children will develop bipolar disorders as they mature [Strober and Carlson, 1982; Kovacs et al., 1994; Kovacs, 1996, 1997; Geller et al., 2001]. We cannot determine at this time if the association signal can be attributed to the portion of the sample that will eventually develop bipolar disorder or if there is a relationship to depressive disorders as well. Family and twin studies indicate bipolar and depressive disorders share substantial genetic overlap [McGuffin et al., 2003] and this locus has also been linked to recurrent depressive disorder [McGuffin et al., 2005], thus this gene may be a contributor more generally to mood disorders. Longitudinal follow up of these children as well as studies of this gene in samples of adults with depressive disorders are required.
Thus far no coding region changes have been identified that can account for the association in bipolar disorder or schizophrenia [Chumakov et al., 2002; Williams et al., 2006]. The one coding region change (rs2391191) that has been genotyped in several studies, Lys30Arg, does not explain the association as a single risk allele in this gene. The arginine allele (frequency 0.627) is over transmitted in our sample as well as in the Williams et al. 2006 study, however not in others [Detera-Wadleigh and McMahon, 2006].
Based on this data, changes in gene expression are predicted and indeed gene expression studies have indicated a tendency toward over expression of G72 but not G30 in the dorsal lateral prefrontal cortex in a post mortem study of brains from individuals with schizophrenia compared to controls [Korostishevsky et al., 2004]. Although the expression of G30 did not differ between the case and control groups, the expression of G72 and G30 was correlated, suggesting a role for G30 in the regulation of G72 [Korostishevsky et al., 2004]. The identification of gene regulatory elements and genetic variation within these elements is thus the next logical step in the study of this gene.
Acknowledgements
The National Institute of Mental Health Program Project grant, MH 56193 and the National Alliance for Research on Schizophrenia and Depression, supported this work.




