Decreased Level of TMEM100 in Neonates With Lethal Lung Developmental Disorders due to Abnormalities in SHH-FOXF1 and TBX4-FGF10 Signaling Pathways
Funding: This work was supported by the National Science Centre in Poland.
ABSTRACT
Lethal lung developmental disorders (LLDDs), histologically classified as alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV), congenital alveolar dysplasia (CAD), acinar dysplasia (AcDys), and primary pulmonary hypoplasia (PH), are rare diseases associated with high neonatal mortality due to refractory respiratory failure. Although ACDMPV mostly results from single nucleotide variants (SNVs) or copy-number variants (CNVs) involving FOXF1, AcDys, CAD, and PH are often associated with abnormalities within TBX4 or FGF10. These genes interact in the SHH-FOXF1 and TBX4-FGF10 signaling network and are known regulators of lung development. Recent studies conducted in TBX4-, FGF10-, or FOXF1-deficient LLDD lungs revealed decreased expression of TMEM100 at the transcriptomic and immunohistochemical levels. Here, we present four new patients with genetically and histopathologically confirmed LLDD, including ACDMPV (n = 2), AcDys (n = 1), and PH (n = 1), in whom we detected a heterozygous variants involving FOXF1 (n = 2) or TBX4 (n = 2). Additional immunohistochemical (TMEM100) and qPCR analyses (TMEM100, TBX4, FOXF1) performed in lung tissues of these newborns revealed a significant reduction in TMEM100, TBX4, and FOXF1 expression. Our results confirm previous findings indicating the possible involvement of TMEM100 in FOXF1-TBX4-FGF10 molecular signaling that, when disrupted, may lead to LLDD.
1 Introduction
Lethal lung developmental disorders (LLDDs) are rare neonatal diseases manifesting in the first hours or days of life as acute respiratory failure with pulmonary arterial hypertension (PAH) that is often refractory to treatment (Vincent et al. 2019). These disorders are associated with very high neonatal mortality (80%–90%), and only a few cases with longer survival have been reported (Ito et al. 2015; Shankar et al. 2006; Szafranski et al. 2019; Vincent et al. 2019). Accurate diagnosis of LLDD is challenging and usually requires clinical and histopathological assessment in combination with genetic testing (Vincent et al. 2019).
Based on histopathological lung features, LLDDs have been classified as acinar dysplasia (AcDys), congenital alveolar dysplasia (CAD), alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV), and primary pulmonary hypoplasia (PH) (Deutsch 2020; Vincent et al. 2019). While ACDMPV is mainly caused by single nucleotide variants (SNVs) or copy-number variant (CNV) deletions involving FOXF1 or its lung-specific enhancer (Stankiewicz et al. 2009; Szafranski, Herrera et al. 2016; Szafranski et al. 2021), AcDys, CAD, or PH are often associated with alterations within the TBX4 or FGF10 genes (Karolak, Vincent et al. 2019; Karolak et al. 2020).
Recent studies of lung tissues derived from patients with FOXF1-, TBX4-, or FGF10-related LLDD revealed downregulation of TMEM100 at the transcript and protein levels (Karolak et al. 2021, 2022; Szafranski et al. 2024). Since TMEM100 is important for vascular development and lung morphogenesis (B. Liu et al. 2022; Moon et al. 2010, 2015; Somekawa et al. 2012), its deficiency may lead to prenatal arrest of lung maturation, resulting in LLDD.
Here, we present four new neonates with LLDD, including ACDMPV (n = 2), AcDys (n = 1), and PH (n = 1) due to a heterozygous variants involving FOXF1 (n = 2) or TBX4 (n = 2), in whom we observed a diminished level of TMEM100 in the lungs.
2 Materials and Methods
2.1 Human Subjects
Peripheral blood samples and/or formalin-fixed paraffin-embedded (FFPE) lung tissues were obtained from four neonates with respiratory failure (LLDD009, LLDD011, LLDD013, LLDD014) and two age-matched control individuals. Saliva samples were collected from the parents of patient LLDD011. Characteristics of all enrolled patients are detailed in Table 1. Samples were collected after informed consent was obtained. The study protocol was approved by the Institutional Review Board for Human Subjects at Poznan University of Medical Sciences.
| ID | Gender | Birth (weeks) | Survival | Molecular findings | Histopathological diagnosis | Immunohistochemistry for TMEM100 |
|---|---|---|---|---|---|---|
| LLDD009 | F | Term | 1D |
TBX4 NM_001321120.2:c.121G>T NP_001308049.1:p.Gly41Ter |
PH Superimposed HMD |
3+ |
| LLDD011 | F | Term | 10D |
FOXF1 NM_001451.3:c.445C>T NP_001442.2:p.Gln149Ter |
ACDMPV | 1+ |
| LLDD013 | M | 38; natural | 5D | ~ 2.2 Mb CNV deletion at 17q23.2 (encompassing TBX4) |
AcDys Superimposed HMD |
2+ |
| LLDD014 | F | 39 | 27D |
FOXF1 NM_001451.3:c.852_856del NP_001442.2: p.Tyr284_Lys286delinsTer |
ACDMPV | 1+ |
- Abbreviations: ACDMPV, alveolar capillary dysplasia with misalignment of pulmonary veins; AcDys, acinar dysplasia; CNV, copy-number variant; D, day; F, female; HMD, hyaline membrane disease; M, male; PH, pulmonary hypoplasia.
2.2 Histopathological Evaluation
Histopathological evaluation was performed using FFPE lung tissue slides obtained at autopsy and stained with hematoxylin and eosin. Immunohistochemical analysis for TMEM100 (1:50, Sigma HPA055936) was carried out on FFPE 5-μm sections on the Ventana BenchMark Ultra stainer after CC1 antigen recovery. Interpretation of TMEM100 immunohistochemistry used the following scoring system: 0+, negative; 1+, rare foci; 2+, scattered foci; 3+, numerous foci; 4+, strong diffuse staining.
2.3 Molecular Analyses
DNA was extracted from peripheral blood samples or FFPE lung tissues using the Gentra Purgene Blood Kit (Qiagen, Hilden, Germany) or the DNeasy Blood & Tissue Kit (Qiagen), respectively. DNA from saliva was isolated using the GeneFix Saliva-Prep DNA Kit (Isohelix, Harrietsham, Kent, United Kingdom). Paired-end (2x150 bp) genome sequencing (GS) was performed in samples LLDD011 and LLDD013 using the NEBNext Ultra II FS DNA Library Prep Kit for Illumina (New England BioLabs, Ipswich, MA) on the NovaSeq 6000 (Illumina, San Diego, CA). Coding regions of FOXF1, TBX4, and FGF10 for patients LLDD009 and LLDD014 were Sanger sequenced with the BigDye Terminator 3.1 kit (Thermo Fisher Scientific, Waltham, MA).
Identified variants were annotated with allele frequencies from gnomAD v4.1.0 (Karczewski et al. 2020), disease associations from ClinVar (Landrum et al. 2014), in silico functional predictions including pathogenicity scores from dbNSFP (Liu et al. 2020), and ACMG variant classification (Richards et al. 2015) using the GeneBe tool (Stawiński and Płoski 2024).
RNA was extracted from FFPE lung tissues obtained from all LLDD patients and normal controls using the Quick-RNA FFPE Kit (Zymo Research, Irvine, CA). Qualitative and quantitative analysis of the isolated RNA was completed using the Qubit RNA (HS) Assay kit (Thermo Fischer Scientific).
qPCR analysis was performed to assess the expression of TMEM100, TBX4, and FOXF1 in lung tissues of LLDD patients and control individuals. RNA samples were reverse transcribed using the GoScript Reverse Transcription kit (Promega, Madison, WI). The qPCR reactions were performed on the LightCycler 96 (Roche, Mannheim, Germany) instrument with FastStart Essential DNA Green Master (Roche) and primers listed in SI Table S1, according to the manufacturer's instructions. All samples were run in triplicates (TMEM100) or duplicates (TBX4 and FOXF1) in three independent experiments. The comparative ΔΔCT method was used to evaluate the relative difference in the number of transcripts. TMEM100, TBX4, and FOXF1 expression values were normalized relative to GAPDH.
Statistical analysis was performed using the one-way ANOVA and post hoc Tukey analyses, in which a p-value less than 0.05 was considered statistically significant.
3 Results
3.1 Case Presentations
Patient LLDD009 was a Caucasian female born at term through cesarean section with a weight of 3730 g and an Apgar score of 10/10. Shortly after birth, she developed respiratory distress, and nasal continuous positive airway pressure (nCPAP) therapy was initiated, followed by intubation and mechanical ventilation. A chest X-ray showed mediastinal emphysema and pneumothorax. During the next few hours, her condition deteriorated, and the patient died on the 2nd day of life.
Patient LLDD011 was a Caucasian female born at 37 weeks of gestation. Apgar scores were 5/1, 7/3, 9/5, and 9/10. Pregnancy was complicated by polyhydramnios. Clinical evaluation revealed pyloric stenosis and cardiac anomalies including atrial septal defect and PAH. Due to the worsening clinical condition, mechanical ventilation and nitric oxide (NO) were initiated within the first day of life with no subsequent appreciable improvement. She died on the 10th day of life.
Patient LLDD013 was a Caucasian male born via vaginal delivery at 38 weeks of gestation with a weight of 3050 g and Apgar scores 5/1, 7/3, 7/5, and 8/10. Minutes after birth, he had respiratory insufficiency with progressive PAH and bilateral pneumothoraces requiring mechanical ventilation. The patient died on the 5th day of life.
Patient LLDD014 was a Caucasian female, born at 39 weeks of gestation weighing 4420 g with Apgar scores of 7/9. The pregnancy was complicated by respiratory infections at 26 and 36 weeks of gestation. The patient was diagnosed with a congenital heart defect (aortic coarctation). She developed PAH shortly after birth. Standard therapy, including high-frequency mechanical ventilation and inhaled NO, was unsuccessful. Extracorporeal membrane oxygenation (ECMO) was started on the first day of life. Despite 25 days of ECMO therapy, there was no clinical improvement. Due to the lack of response to therapy and a suspected diagnosis of LLDD, ECMO was discontinued. The patient expired on the 27th day of life.
3.2 Histopathological Findings
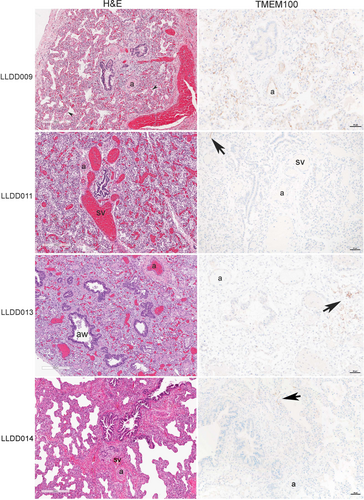
On histopathological evaluation, lung sections from patient LLDD009 demonstrated PH with immature architecture for term gestation and superimposed hyaline membranes. Arterial thrombi were also seen. Immunohistochemical expression for TMEM100 was present within the endothelium of nearly all arteries and lobular capillaries but was generally weak.
Lung sections from patient LLDD011 revealed abnormal dilated shunt vessels (“misaligned pulmonay veins”) adjacent to airways and arteries characteristic of ACDMPV. Arteries showed hypertensive changes with medial hypertrophy and the alveolar architecture was simplified. TMEM100 protein level was markedly reduced with only weak expression seen within a rare vessel.
Autopsy lung from patient LLDD013 demonstrated the classic findings of AcDys. There was arrested lung development with only bronchioles and simplified acinar structures present, the latter containing superimposed hyaline membrane material and inflammatory debris. TMEM100 expression was limited to scattered arteries and foci of capillaries.
Lung sections from patient LLDD014 demonstrated ACDMPV with abnormally dilated shunt vessels in the bronchovascular bundle and hypertensive changes of the pulmonary arteries. TMEM100 expression was limited to rare foci of capillaries.
Hematoxylin and eosin staining and immunohistochemistry for TMEM100 sections for all patients are presented in Figure 1.
3.3 Molecular Findings
3.3.1 Genome Sequencing and/or Sanger Sequencing
Genetic analysis in patient LLDD009 revealed a heterozygous c.121G>T (p.Gly41*) variant in TBX4 (NM_001321120.2, NP_001308049.1) that is absent in the gnomAD (v4.1.0) database (Table 1, SI Figure S1). This variant is predicted by in silico tools to be pathogenic due to the premature termination of a protein and is expected to be a subject to degradation by nonsense-mediated decay (NMD). According to ACMG guidelines, the c.121G>T variant in TBX4 is classified as pathogenic (PVS1 very strong, PM2 moderate, PP5 supporting; score 11), and as likely pathogenic by the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar, Version: 05-Dec-2024). Parents were not available for testing.
In proband LLDD011, trio-based genetic testing revealed a de novo heterozygous c.445C>T (p.Gln149*) variant in FOXF1 (NM_001451.3, NP_001442.2) (Table 1, SI Figure S2). This variant is not present in the control population dataset and results in NMD. The pathogenic outcome for the c.445C>T variant in FOXF1 is predicted by in silico tools. Using ACMG criteria, the variant is predicted to be pathogenic (PVS1 very strong, PS2 strong, PM2 moderate; score 14).
Using GS, a heterozygous recurrent ~2.2 Mb CNV deletion at 17q23.1q23.2 (~chr17:60,035,000-62,265,000, hg38) encompassing TBX4 and TBX2 was detected in patient LLDD013 (Table 1, SI Figure S3). Parents were not available for testing.
Genetic analysis in patient LLDD014 revealed a heterozygous c.852_856del (p.Tyr284*) variant in FOXF1, leading to a premature termination of the protein (Table 1, SI Figure S4). Its deleterious effect is predicted by in silico functional annotation. The variant is absent in the gnomAD database and is a subject to degradation by NMD. Using ACMG criteria, the variant is classified as pathogenic (PVS1 very strong, PM2 moderate, PP5 moderate; score 12), as well as by the ClinVar database (Version: 05-Dec-2024). Parents were not tested.
3.3.2 qPCR and Statistical Analysis
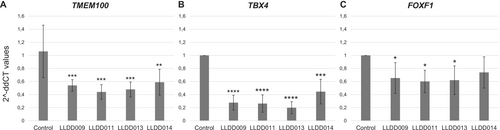
qPCR analysis performed in lung tissues of four newborns affected by LLDD showed a statistically significant decrease in the number of TMEM100, TBX4, and FOXF1 transcripts compared to normal lungs derived from age-matched individuals (Figure 2A). The highest reduction of TMEM100 expression was observed in newborn LLDD011 (58.5%, p = 0.000194), while the lowest reduction was detected in patient LLDD014 (44.5%, p = 0.002366). In patients LLDD009 and LLDD013, the TMEM100 level was decreased by 49.2% (p = 0.000876) and 54.8% (p = 0.000319), respectively. TBX4 levels were statistically significantly reduced in all patients, with the highest decrease observed in patient LLDD013 (80.2%, p = 0.000283) and the lowest reduction detected in newborn LLDD014 (55.5%, p = 0.002013). In patients LLDD009 and LLDD011, levels of TBX4 transcripts were decreased by 72.4% (p = 0.000439) and 73.8% (p = 0.000397), respectively (Figure 2B). The highest reduction of FOXF1 expression was observed in patient LLDD011 (39.9%, p = 0.020079). In patients LLDD009 and LLDD013, the FOXF1 level was decreased by 34.7% (p = 0.041283) and 37.9% (p = 0.026206), respectively. In newborn LLDD014, the reduction of FOXF1 was observed (26.1%, p = 0.139834), but it was not statistically significant (Figure 2C).
4 Discussion
Approximately 180 SNVs and CNV deletions encompassing FOXF1 have been described in ACDMPV patients to date, representing ~90% of all ACDMPV cases (Bzdęga et al. 2023; Stankiewicz et al. 2009; Szafranski, Gambin et al. 2016; Szafranski, Herrera et al. 2016; Szafranski et al. 2019; Yıldız Bölükbaşı, Karolak, Gambin et al. 2022). In contrast, 18 SNVs or CNVs involving TBX4 (Bzdęga et al. 2024; Karolak et al. 2020; Karolak, Szafranski et al. 2019; Karolak, Vincent et al. 2019; Suhrie et al. 2019; Szafranski, Coban-Akdemir et al. 2016; Szafranski et al. 2024, 2025; Yıldız Bölükbaşı, Karolak, Szafranski et al. 2022) and 9 SNVs or CNVs encompassing FGF10 (Karolak, Vincent et al. 2019; Schütz et al. 2023; Wade et al. 2022) have been reported in neonates with other subtypes of LLDD, including AcDys, CAD, and primary PH, accounting for ~45% and ~20% of these cases, respectively. Here, we describe two novel patients with genetic and histopathological diagnoses of ACDMPV and one patient each with AcDys and PH.
We identified a de novo heterozygous c.445C>T variant (p.Gln149*) in FOXF1 in patient LLDD011 with ACDMPV, which is also expected to cause premature termination of the FOXF1 protein. This variant is novel and has not been previously reported in association with ACDMPV or other diseases. In another newborn with ACDMPV, patient LLDD014, we detected a heterozygous c.852_856del variant (p.Tyr284*) in FOXF1. Previously, the same variant was reported in one patient with ACDMPV, but no phenotypic or clinical specifics are available for this case (VCV001302014.2—ClinVar—NCBI n.d.).
In patient LLDD009 with PH, we detected a heterozygous stop gain c.121G>T (p.Gly41*) variant in TBX4. Interestingly, the same variant was previously identified in the patient from the UK PAH cohort (Gräf et al. 2018). In patient LLDD013 with AcDys, we found a heterozygous recurrent ~2.2 Mb deletion at 17q23.1q23.2, involving TBX4, that was earlier reported in a few patients with LLDD (Karolak, Vincent et al. 2019), as well as patients with various congenital defects including ischiocoxopodopatellar syndrome with or without PAH (Galambos et al. 2019; Kerstjens-Frederikse et al. 2013) or developmental delay, microcephaly, heart defects, limb abnormalities, and hearing loss (Ballif et al. 2010). Heterogeneity in the clinical features observed in individuals with the same TBX4 variant suggests that those changes alone are insufficient to cause the specific phenotypes and additional modifiers are required (Karolak et al. 2023). Recently, a complex compound inheritance model of coding and noncoding variants within the TBX4 locus has been proposed to explain variable phenotypic expressivity (Karolak, Vincent et al. 2019; Yıldız Bölükbaşı, Karolak, Szafranski et al. 2022).
FOXF1, TBX4, and FGF10 are members of the FOXF1-TBX4-FGF10 signaling pathway and their expression in developing lungs is controlled by SHH (Fernandes-Silva et al. 2017). SHH signals from epithelial cells activate Foxf1 and Tbx4 in the lung mesenchyme (Mahlapuu et al. 2001). As a result, activated transcription factors work in the complex network, mediating expression of Fgf10 in the lung mesenchyme to ensure proper branching morphogenesis (Fernandes-Silva et al. 2017; Karolak et al. 2021, 2023). While the above data indicate that complex epithelial-mesenchymal signaling through the SHH-FOXF1 and TBX4-FGF10 is crucial for lung development, the exact interactions between these and other factors involved in lung morphogenesis remain not fully understood.
Recent studies revealed that TMEM100 may also play a fundamental role in lung development (Liu et al. 2022). TMEM100 is highly expressed in human fetal lungs and acts downstream of the BMP9/BMP10-ALK1 signaling pathway (Pan et al. 2023). In Tmem100 −/− mice, embryonic death has been observed due to severely impaired vascular morphogenesis (Somekawa et al. 2012), indicating its role in regulating blood vessel and early lung development (Somekawa et al. 2012). Interestingly, an earlier ChiP-seq study performed in mice with Foxf1 overexpression showed that Tmem100 may be a target of FOXF1 (Dharmadhikari et al. 2016). Recent transcriptomic and immunohistochemical studies conducted in lung tissues of newborns with LLDD due to FOXF1, TBX4, or FGF10 deficiency noted reduced expression of TMEM100 in all patients, suggesting potential interaction between TMEM100 and the FOXF1-TBX4-FGF10 gene network and implying that disruption of this crosstalk may be important in the pathogenesis of LLDD (Karolak et al. 2021, 2022). Decreased TMEM100 at the transcriptomic and proteomic levels was also later observed in another neonate with AcDys caused by a missense c.728A>C (p.Asn243Thr) variant in TBX4 (Szafranski et al. 2024) and a newborn with CAD associated with a ~1.07 Mb heterozygous CNV deletion at 17q23.2 involving TBX4 (Bzdęga et al. 2024).
Our study involving four neonates with FOXF1- and TBX4-related LLDD also showed significantly decreased TMEM100 expression on the transcript level compared to control lungs obtained from age-matched neonates without LLDD, confirming previous findings. In patients LLDD011 and LLDD013, reduced TMEM100 expression observed on the gene level correlates with decreased protein level identified by immunohistochemistry. However, in patient LLDD009, the expression of TMEM100 was higher on the protein level than at the gene level. Since we used the tissues obtained postmortem and fixed with formalin and paraffin, we cannot exclude that RNA in this sample was partially degraded disturbing the ability to properly estimate the expression level. Variability in TMEM100 expression can be seen in tissues with autolysis and/or poor fixation (G. Deutsch, personal observation).
In addition to downregulation of TMEM100, our analysis also showed a reduced level of TBX4 and FOXF1 transcripts in the lungs of all studied patients with both TBX4- or FOXF1-related abnormalities, further confirming molecular crosstalk between TMEM100 and the FOXF1-TBX4-FGF10 network (Karolak et al. 2021, 2022) during human lung development.
In summary, we present clinical, histopathological, and molecular findings in four newborns with LLDD. The identification of genetic variants in TBX4 or FOXF1 in additional patients allows us to expand the genotypic spectrum of LLDD. Marked diminished expression of TMEM100 at both gene and protein levels, as well as decreased expression of the TBX4 and FOXF1 genes, observed in our patients with ACDMPV, AcDys, or PH associated with FOXF1 or TBX4 variants, further implies potential interaction between TMEM100 and FOXF1–TBX4–FGF10 signaling and supports the hypothesis that deficiency of TMEM100 plays an important role in LLDDs pathogenesis. Our study suggests that decreased levels of TMEM100 could be useful in the LLDD diagnosis.
Author Contributions
Katarzyna Bzdęga: formal analysis, investigation, methodology, validation, visualization, writing – original draft, writing – review and editing. Gail H. Deutsch: formal analysis, investigation, methodology, visualization, writing – original draft, writing – review and editing. Małgorzata Rydzanicz: investigation, methodology, writing – review and editing. Witold Błaż: formal analysis, investigation, writing – review and editing. Elżbieta Rafińska-Ważny: formal analysis, investigation, writing – review and editing. Anna P. Terpin: formal analysis, investigation, writing – review and editing. Dariia Klepach: formal analysis, investigation, writing – review and editing. Valentyna Zakharova: formal analysis, investigation, writing – review and editing. Rafał Płoski: investigation, methodology, writing – review and editing. Tomasz Szczapa: formal analysis, investigation, writing – review and editing. Justyna A. Karolak: conceptualization, formal analysis, ivestigation, methodology, validation, visualization, project administration, writing – original draft, writing – review and editing.
Acknowledgments
This research was supported by the National Science Centre in Poland grant 2019/35/D/NZ5/02896 (J.A.K.) and Poznan University of Medical Sciences Doctoral School grant number NMN0000084 for 2023 (K.B.) financed from the statutory funds. Genome sequencing was performed thanks to Genomics Core Facility CeNT UW, using the NovaSeq 6000 platform financed by the Polish Ministry of Science and Higher Education (decision no. 6817/IA/SP/2018 of 2018-04-10). We thank the families whose help and participation made this work possible.
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
The data that supports the findings of this study are available in the Supporting Information of this article.




