Known pathogenic gene variants and new candidates detected in sudden unexpected infant death using whole genome sequencing
Abstract
The purpose of this study is to gain insights into potential genetic factors contributing to the infant's vulnerability to Sudden Unexpected Infant Death (SUID). Whole Genome Sequencing (WGS) was performed on 144 infants that succumbed to SUID, and 573 healthy adults. Variants were filtered by gnomAD allele frequencies and predictions of functional consequences. Variants of interest were identified in 88 genes, in 64.6% of our cohort. Seventy-three of these have been previously associated with SIDS/SUID/SUDP. Forty-three can be characterized as cardiac genes and are related to cardiomyopathies, arrhythmias, and other conditions. Variants in 22 genes were associated with neurologic functions. Variants were also found in 13 genes reported to be pathogenic for various systemic disorders and in two genes associated with immunological function. Variants in eight genes are implicated in the response to hypoxia and the regulation of reactive oxygen species (ROS) and have not been previously described in SIDS/SUID/SUDP. Seventy-two infants met the triple risk hypothesis criteria. Our study confirms and further expands the list of genetic variants associated with SUID. The abundance of genes associated with heart disease and the discovery of variants associated with the redox metabolism have important mechanistic implications for the pathophysiology of SUID.
1 INTRODUCTION
Sudden Infant Death Syndrome (SIDS) is defined as the sudden and unexpected death of an infant younger than one year, for which the cause of death remains unexplained despite a thorough investigation including a complete autopsy, and review of the circumstances of death along with review of the clinical history (American SIDS Institute, 2023). The term Sudden Unexpected Infant Death (SUID) is more encompassing. Using the International Classification of Diseases, 10th Revision (ICD-10), both the Centers for Disease Control and Prevention (CDC) and the American Academy of Pediatrics (AAP) define SUID as a larger category that encompasses three types of infant death in children prior to one year of age: “sudden infant death syndrome (SIDS; R95), deaths from other ill-defined or unknown causes (R99), and accidental suffocation and strangulation in bed (W75)” (Lavista Ferres et al., 2020). SUID remains a significant public health problem. One of the leading causes of infant mortality, it is estimated that SUID contributes to the deaths of approximately 3400 infants in the United States each year (Centers for Disease Control and Prevention, 2023). The use of these certified causes of death has varied over time and varies by jurisdiction (Goldstein et al., 2019).
Many risk factors have been identified for SIDS/SUID including race/ethnicity (Centers for Disease Control and Prevention, 2023), prematurity (Allen et al., 2021), maternal smoking during pregnancy (Allen et al., 2021; Anderson et al., 2019), lower birth weight (Allen et al., 2021), bed-sharing (Allen et al., 2021), prone sleeping (Mitchell, 1991; Mitchell & Krous, 2015), recent infection (Goldwater, 2017), geographic variations (Mitchell et al., 2020), and environmental exposures such as air pollution (Dales et al., 2004). The recognition of prone sleeping as a major risk factor led to the adoption of safe sleep practices which was associated with significant reductions in SUID rates in the 1990's and early 2000's. Increased educational efforts (Mitchell, 1997) could further increase the adherence to safe sleep practices, and also reduce the prevalence of maternal smoking, which in turn could decrease the number of SUID deaths. Unfortunately, despite increased educational efforts, SUID rates have plateaued since the early 2000's (Centers for Disease Control and Prevention, 2023).
Further deaths might be prevented through the early recognition of a child that is at higher risk for SUID. DNA sequencing has identified genes related to cardiac, neurologic, and metabolic disorders that could be linked to SUID (Baruteau et al., 2017; Chahal et al., 2020; Chahal et al., 2021; Clemens et al., 2020; Gray et al., 2019; Halvorsen et al., 2021; Keywan et al., 2021; Koh et al., 2022; Männikkö et al., 2018; Moore et al., 2020; Opdal, 2018; Rochtus et al., 2020; Tester, Wong, Chanana, Gray, et al., 2018; Tester, Wong, Chanana, Jaye, et al., 2018). The early identification of high-risk genes could potentially also be important for preventing mortality and morbidity for children that survive beyond the one-year window for SUID. Identifying risk genes could also help in the diagnosis of a sudden death and bring closure to surviving members of a family who lost a child. With the aid of genetic counseling, parents could be better informed of the risk of recurrence when considering further children. The knowledge of a genetic vulnerability together with improved recognition devices that monitor multiple physiological parameters, could also increase the usefulness of monitoring equipment that have so far failed to prevent deaths (Ramanathan et al., 2001).
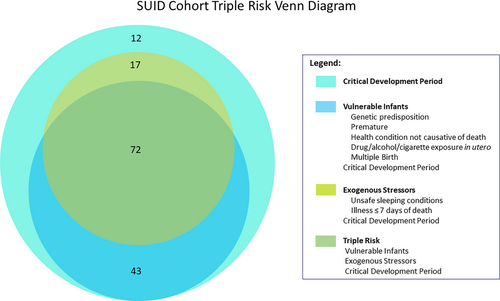
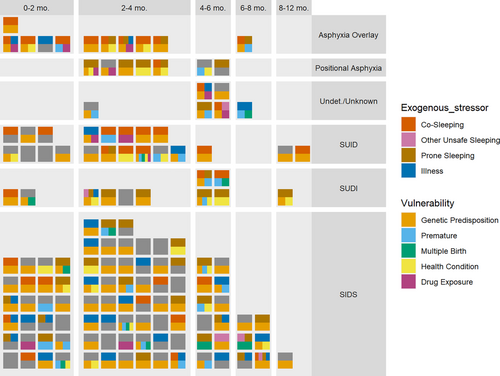
The triple risk model provides an important conceptual framework to assess how a potentially pathogenic gene variant could contribute to a fatal outcome (Guntheroth & Spiers, 2002). First proposed in 1972, but evolved over time, this concept puts forth the hypothesis that SUID occurs during a critical development period when a vulnerable infant is exposed to an extrinsic stressor (Guntheroth & Spiers, 2002; Rognum & Saugstad, 1993). A pathogenic gene variant could interfere with any of the three risk factors. It could increase or imbue a child with an intrinsic vulnerability, it could interfere with the response to a particular external stressor, and it could alter the normal developmental trajectory. Importantly, it is generally assumed that none of these isolated threats are significant enough to cause death alone, but when combined, the triple risks reach the threshold for a fatal outcome. Based on numerous neuropathological and neurophysiological studies it is hypothesized that the concurrence of the triple risk (developmental window of post-perinatal age (day 6–364), intrinsic vulnerability and external stressor) leads in most cases to failed arousal in response to external stressors associated with hypercapnic and/or hypoxic conditions (Garcia 3rd et al., 2013; Kinney & Haynes, 2019; Nasirova et al., 2020; Vivekanandarajah et al., 2021). The conditions become fatal as the child suffers a terminal apnea event leading to severe irreversible hypoxic damage and ultimately cardiac arrest due to the compromised arousal response. However, there are also reports that a child can succumb to apnea, sudden cardiac arrest or perhaps a catastrophic immunological response, such as a cytokine storm, that is not caused by a failed arousal (Opdal & Rognum, 2011; Opdal, 2018). On the other hand, a slight upper airway infection in combination with the prone position may affect arousal and increase the risk for SIDS in a synergistic manner (Ferrante et al., 2024). Risk genes associated with cardiac and neurologic disorders including cardiomyopathy and channelopathies, metabolic conditions such as fatty acid oxidation or glucose metabolism errors, or altered immune responses and inflammation (Baruteau et al., 2017; Chahal et al., 2020; Chahal et al., 2021; Clemens et al., 2020; Gray et al., 2019; Halvorsen et al., 2021; Keywan et al., 2021; Koh et al., 2022; Männikkö et al., 2018; Moore et al., 2020; Opdal, 2018; Rochtus et al., 2020; Tester, Wong, Chanana, Gray, et al., 2018; Tester, Wong, Chanana, Jaye, et al., 2018) could be potential candidates for such fatal events. While these considerations support the notion that a multitude of pathways can lead to SUID, there are certain features of SUID that are shared and are reproduced world-wide. These common features include male dominance (Allen et al., 2021), the characteristic age distribution which peaks at the second month and ends at one year of age (Allen et al., 2021) and maternal smoking (Anderson et al., 2019) as well as prone sleeping (Mitchell, 1991; Mitchell & Krous, 2015).
Next-generation sequencing (NGS) studies have accelerated the study of a wide range of heritable diseases. In the present study, we performed WGS of 144 children that succumbed to SUID and categorized them using the triple risk hypothesis as a framework to measure their risk for this tragic event (Figure 1). Our goals were to (1) validate genes in which altered function have been previously implicated in SUID and (2) identify novel genes and pathways that tend to be disrupted in SUID so that we can better understand the causes of these deaths. Our complete, nontargeted approach to sequencing enables an unbiased and thorough evaluation of the SUID genome.
2 MATERIALS AND METHODS
2.1 Editorial policies and ethical considerations
Samples obtained from the NIH's NeuroBioBank (NBB) (https://neurobiobank.nih.gov/) were exempt from the requirement of informed consent because the individuals providing the samples were deceased. However, donor recruitment sites for the NIH NBB typically obtain authorization from the parent/s of the individual. Archived samples obtained from the University of Washington and Seattle Children's Hospital were granted a waiver for informed consent through Seattle Children's Research Institute's Institutional Review Board (IRB), which approved the study. Informed consent was obtained for samples which did not meet criteria for the waiver granted by the IRB. This study is Health Insurance Portability and Accountability Act (HIPAA), General Data Protection Regulation (GDPR), and NIH compliant.
2.2 Affected infant/healthy adult acquisition
2.2.1 Affected infants
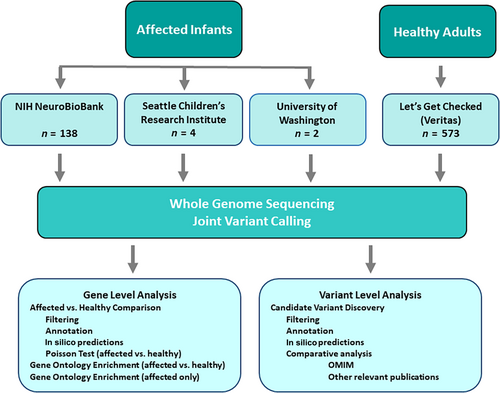
Source of material: Samples from infants lost to SUID were obtained from three sources: 1. The NIH NBB (n = 140, year of death from 1992 to 2017), 2. Seattle Children's Research Institute (SCRI) (n = 4, year of death from 2010 to 2014), and 3. University of Washington (UW) (n = 2, year of death was 2008).
Affected infant definition: Autopsy reports and partial medical history were available for only 34 infants, but for 93 infants, some medical examiner (ME) notes were included. For the remainder of the cohort, limited data were provided. The cause of death as stated by the ME was accepted. Deaths where there was clear mechanical asphyxia were excluded (n = 2). Cause of death of the 144 infants included in the study were SIDS n = 88, unexplained/sudden unexplained infant death n = 21, undetermined/unknown n = 6, sudden unexpected infant death n = 11, positional asphyxia n = 7, asphyxia/overlay n = 11.
Postmortem tissue: Tissue was obtained from the 146 infants. For the majority this was brain (96.5%), but for some infants, only liver tissue or blood was available.
2.2.2 Healthy adults
Sequencing results (Variant Call Format files (VCFs) from 573 healthy adults were obtained from Veritas (newly acquired by Let's Get Checked). The control population ranged in age from 18 to 100+, with 45 (7.85%) over 100 years old (Let's Get Checked, 2023).
See Figure 2 for flow chart.
2.3 Whole genome sequencing
All affected infant and healthy adult samples were subjected to a quality control assessment for high molecular weight DNA and degradation utilizing SNP chip and TapeStation prior to library construction (Let's Get Checked, 2023). Samples that passed the quality assessment were processed with the TruSeq DNA PCR-free sample preparation kit and sequenced (Let's Get Checked, 2023). Samples were sequenced to an average of approximately 30× coverage on the Illumina HiSeq × 10 or NovaSeq 6000 next generation sequencers (Let's Get Checked, 2023). The paired-end sequencing protocol was used targeting an average read-length of 150 base pairs (Let's Get Checked, 2023). Samples were required to meet or exceed 97% of bases >10× coverage (Let's Get Checked, 2023). Veritas performed primary analysis on the results which yielded two Fast Quality Score files (FASTQs) per sample. Secondary analysis was also performed by Veritas using Microsoft Genomics Service and yielded one Binary Alignment/Map (BAM) file and genomic variant call file (gVCF) per sample. Tertiary analysis was completed at Microsoft Azure Data Science Virtual Machine (MSFT-AZURE-DSVM) and SCRI. Joint genotype calling was performed with GATK GenotypeGVCFs on MSFT-AZURE-DSVM.
2.4 Population structure analysis
To confirm a lack of major batch effects between affected infants and healthy adults, population structure was visualized across the 717 individuals. The joint-called VCF was pruned with BCFtools (Li, 2009) to only retain variants with less than 10% missing data, removing variants with linkage disequilibrium of 0.25 or greater with another variant, retaining a maximum of 10 variants per 100 kb. The variants were then imported into R and further filtered to only retain autosomal, biallelic variants with excess heterozygosity below 125 and an alternative allele frequency between 0.02 and 0.5, yielding 9252 variants. Principal components analysis, using the PPCA method from the pcaMethods Bioconductor package to handle missing data, was run on the genotype matrix (Stacklies et al., 2007). The first 50 axes were retained and used as input to UMAP (McInnes et al., 2018) for visualization. Self-identified race and results from GrafPop (Jin et al., 2019) were used to determine the ancestry of each cluster (Figure 3).

2.5 Gene level analysis
2.5.1 Annotation and filtering
BCFtools (Li, 2009), SnpEff (Cingolani, Platts, et al., 2012), Slivar (Pedersen et al., 2021), ANNOVAR (Wang et al., 2010), and SnpSift (Cingolani, Patel, et al., 2012) were used to add functional annotation, predictions of pathogenicity, gnomAD allele frequencies (Karczewski et al., 2020), and data from ClinVar (Landrum et al., 2014) to the VCF. Variants were excluded if they were listed as benign or likely benign in ClinVar, if their inbreeding coefficient was below −0.4 or above 0.4, if their excess heterozygosity was above 125, or if the alternative allele frequency was greater than 0.05 within the dataset. To eliminate variants with dramatically different call rates between affected and healthy, Fisher's exact test was performed for each variant on the contingency matrix of missing versus called genotypes in affected and healthy, and variants were excluded if the odds ratio was greater than 8 or less than 0.125 and the p-value was less than 0.01. An allele frequency cutoff for maximum frequency in gnomAD was set to 0.0001 for putative dominant variants, and 0.005 for putative recessive variants. Two allele frequency filters were then applied. (1) In the first filter, if a variant was homozygous in any individual, it had to pass the recessive allele frequency threshold in all populations. Other variants had to pass the dominant allele frequency threshold in all populations. This filter yielded 18,401 variants. (2) The second filter was the same as the first, except that variants had to be completely absent in the healthy adults (unless they were homozygotes in the affected and only heterozygotes in the healthy), yielding 2758 variants.
Three filters, decreasing in stringency, were then applied to these two filtered datasets. (1) The first filter retained only variants labeled as Pathogenic (P) or Likely pathogenic (LP) in ClinVar, yielding 119 variants across 102 genes out of the 18,401 variants in affected infants and healthy adults, and 21 variants across 22 genes out of the 2758 variants only in affected infants. (2) The second filter included all variants from the first, plus any that were indicated as deleterious using three out of six computational methods, including MetaSVM, FATHMM or fathmm_MKL_coding, Polyphen2 HDIV, or HVAR, CADD, GERP++, and DANN. This filter yielded 3443 variants across 2667 genes out of the 18,401 variants in affected and healthy, and 655 variants across 632 genes out of the 2758 variants only in affected infants. (3) The third filter included all variants from the first two plus any splicing, frameshift, or stop gain variants. This filter yielded 9717 variants across 5048 genes out of the 18,401 variants in affected and healthy, and 1150 variants across 998 genes out of the 2758 variants only in affected infants.
2.5.2 Gene-level comparison of affected infants
2.5.3 Gene ontology enrichment in affected infants versus healthy adults
A Poisson test similar to that used to detect significant genes was run on 11,793 gene ontology (GO) terms, using the sum of observation of variants across genes within each ontology term. Multiple testing correction was not appropriate given overlap among ontology terms, so instead a p-value cutoff of 0.00001 was used.
2.5.4 Gene ontology enrichment in affected infants only
Lists of genes in affected infants only for each of the three sets were analyzed in STRING-DB for protein–protein interactions and enrichment in GO terms (Szklarczyk et al., 2019). Genes from the three sets were analyzed using the full STRING network. The required score was set to the medium confidence level of 0.400 and FDR stringency was set to medium, 5%.
2.6 Variant level analysis: Gene discovery Mendelian pipeline
The control VCF was decomposed, subset and annotated with an Ensembl gene annotation using bcftools (Let's Get Checked, 2023). Slivar was used to prioritize variants based on quality, coverage, allele balance and population frequency in gnomAD (Karczewski et al., 2020; Pedersen et al., 2021). In addition, Slivar was used to add healthy adult counts and gene-based information for probability of being loss-of-function intolerant (pLI) score and any Online Mendelian Inheritance in Man (OMIM) associations (Hamosh et al., 2022; Pedersen et al., 2021). Annovar was used to add additional variant-based annotation including: refGene model, ClinVar, CADD, GERP++, PolyPhen2 score, and Geno2MP information (Wang et al., 2010).
Variants were prioritized using the following criteria: ≥10 reads; genotype quality ≥15; allele balance of >0.8 for homozygous alternate and between 0.2 and 0.8 for heterozygous; max gnomAD population frequency of ≤1%, and were determined to be functional (including missense, frameshift, or splicing [splice acceptor, splice donor or splice region, i.e., within 8 bp of an exon]). Variants that appeared in healthy adults were excluded.
Included here are variants only described as rare/ultra-rare (<0.005%) in gnomAD, with noted exceptions, associated with autosomal dominant (AD) conditions, though some variants are also associated with multiple conditions including those of autosomal recessive (AR) inheritance. Infants were all heterozygous for the variant unless otherwise stated. When a variant is associated with AR conditions alone, it is only included if the infant had two variant alleles for that gene in different phases. The allele frequency of these variants is reported specific to the race/ethnicity of the infant. When the race/ethnicity of the infant was not available, the total allele frequency for the combined racial/ethnic groups in gnomAD is reported (Karczewski et al., 2020).
3 RESULTS
3.1 Cohort Characteristics
The characteristics were similar to that reported previously (Centers for Disease Control and Prevention, 2023). 50.3% of infants were white and 44.4% were black. 54.2% were boys. The median age at death was 85 days (IQR = 59, 110). There were 10 twins and 1 triplet (Table 1).
| Demographics | Number of cases | Proportion |
|---|---|---|
| Ethnicity/Race | ||
| White | 72 | 50.0% |
| Black or African American | 64 | 44.4% |
| Hispanic | 3 | 2.1% |
| American Indian | 1 | 0.7% |
| Not specified | 4 | 2.8% |
| Sex | ||
| Male | 78 | 54.2% |
| Female | 66 | 45.8% |
| Age at death | ||
| <60 days | 39 | 27.1% |
| 60–180 days | 93 | 64.6% |
| 181–365 days | 12 | 8.3% |
| Multiple birth | ||
| Singlet | 133 | 92.4% |
| Twin | 10 | 6.9% |
| Triplet | 1 | 0.7% |
As the available data were incomplete, it is difficult to report the true frequency of other factors. However, there were 30 infants, 20.8% of the cohort, reported to have had mild illness at or within one week of death. Mostly these illnesses related to the respiratory tract, however, these were not determined to be the cause of the death. Forty-one (28.5% of the cohort) were reported to be co-sleeping with parents and/or siblings, however, for 38 (26.4% of the cohort) it was not known whether they were co-sleeping.
Infants were determined to be vulnerable if they had at least one of the following risk factors: prematurity, genetic predisposition (based on WGS results), exposure to cigarettes, alcohol and/or drugs in utero, having a health condition not determined to be causative of death, or being a twin or triplet. One hundred fifteen (79.9% of the cohort) met these criteria. Eighty-nine infants (61.8%) were described as experiencing an exogenous stressor at or near their time of death. The following risk factors were considered to be exogenous stressors in this study: illness within seven days of death, co-sleeping, prone sleeping, or other unsafe sleep such as excess bedding. All infants in the study died during the critical development period, between one week and one year of age. Risk factors among our cohort as they occur within the triple risk hypothesis categories, vulnerable infant, exogenous stressor, and critical development period are illustrated in Figure 1. Many infants had more than one risk factor in multiple categories, with no clear association between vulnerabilities, exogenous stressors, and age at death (Figure 6).
3.2 Population structure analysis
All major clusters contained both affected and healthy individuals, indicating a lack of confounding batch effects, although ancestry proportions differed between affected and healthy as expected, with affected infants having a much higher proportion of African-Americans (Figure 3). Based on clustering in UMAP, self-reporting of race, and ancestry inference using GrafPop, individuals were divided into three broad ancestry groups: Asian (one affected infant and 35 healthy adults), African (68 affected and 13 healthy), and European (75 affected and 525 healthy).
3.3 Gene level analysis
3.3.1 Genes significantly enriched for functional variants in affected infants versus healthy adults
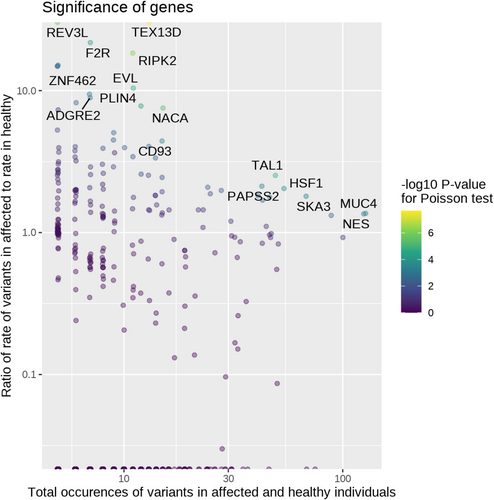
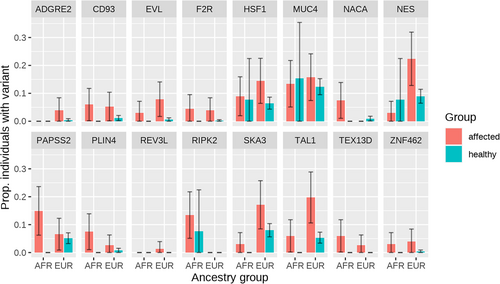
Using 9717 variants that passed the least stringent filtering criteria (P/LP on ClinVar, predicted pathogenic with at least three computational methods, and/or a frameshift, splice site, or stop gain variant), 16 genes were significant at FDR < 0.05 for having a higher affected:healthy ratio for rate of variants than the baseline ratio of 0.647 (Figure 4, Table S1). For some of these genes, affected infants were more likely to have a variant than healthy adults regardless of ancestry, whereas others only showed potentially pathogenic variants in affected infants of European or African ancestry (Figure 5). Significant genes were largely driven by frameshift and splicing variants that were more common in affected than healthy (Table S2).
3.3.2 Gene ontology enrichment in affected infants versus healthy adults
Using the same 9717 variants from the gene-level analysis, 33 gene ontology terms had p < 0.00001 for the affected:healthy ratio of rate of variants exceeding the baseline ratio of 0.647. Out of these 33 terms, 17 had lower p-values than any of their constituent genes (Table S3). Notable terms among those 17 include Golgi lumen (GO:0005796) and maintenance of gastrointestinal epithelium (GO:0030277) driven by variants found across multiple mucin genes; positive regulation of cysteine-type endopeptidase activity involved in apoptotic process (GO:0043280) driven by F2R and HSF1; positive regulation of mitotic cell cycle (GO:0045931) driven by TAL1 and HSF1; and several involving RIPK2 including JUN kinase kinase kinase activity (GO:0004706), response to exogenous dsRNA (GO:0043330), positive regulation of T-helper 1 cell differentiation (GO:0045627), response to interleukin-1 (GO:0070555), response to interleukin-12 (GO:0070671), and cellular response to muramyl dipeptide (GO:0071225).
3.3.3 Gene ontology enrichment in affected infants only
Submission of the first set of 21 genes to STRING which were found with variants in affected infants only, and previously reported as P or LP in ClinVar revealed no significant enrichment in any GO terms (Benjamini et al., 2001). Submission of the second set of 632 genes that included the previous 21 genes in addition to genes containing variants predicted to be deleterious by at least 50% of the computational methods applied which revealed significant enrichment in 142 GO terms at FDR < 0.05 (including 16 GO terms at FDR ≤ 0.0001), with a protein-protein interaction p-value <1.0e−16 indicating more connectivity in the network than would be expected from a random set of genes (Table S4) (Benjamini et al., 2001). Of the 16 most significantly enriched terms, notable are microfilament motor activity (GO:0000146) by a factor of 9, cytoskeletal motor activity (GO:0003774), extracellular matrix structural constituent (GO:0005201) and actin filament binding (GO:0051015) all by a factor of 5, ATP-dependent activity (GO:0140657) by a factor of 3, and protein-containing complex binding (GO:0044877) by a factor of 2. Twenty-four terms were enriched less significantly (FDR <0.001). Of these, four were found to exceed the expected occurrence by a factor of 3: basal plasma membrane (GO:0009925), basal part of cell (GO:0045178), ATP hydrolysis activity (GO:0016887), and actin binding (GO:0003779). Additionally, 44 GO terms were enriched at FDR ≤ 0.01. Most notable among them, minus-end-directed microtubule motor activity (GO:0008569) by a factor of 10, axonemal dynein complex (GO:0005858) and intracellular ligand-gated ion channel activity (GO:0005217) both by a factor of 9, mysoin comlex (GO:0016459) and stereocilium (GO:0032420) by a factor of 6, cardiac cell development (GO:0055006), and cardiac muscle cell differentiation (GO:0055007) both by a factor of 5. Another 58 GO terms were enriched at FDR ≤ 0.05, including ryanodine-sensitive calcium-release channel activity (GO:0005219) by a factor of 32, collagen-activated tyrosine kinase receptor signaling pathway (GO:0038063) by a factor of 13, collagen-activated signaling pathway (GO:0038065) by a factor of 11, and calcium-release channel activity (GO:0015278) by a factor of 10. See Table S4 for a complete list of 142 significantly enriched GO terms in this set of 632 genes.
Submission of the third set of 998 genes to STRING which were found with variants in affected infants only, previously reported as P/LP in ClinVar, predicted by our computational methods to be deleterious and including any frame shift, stop-gained or splicing variants revealed significant enrichment in 78 GO terms at FDR < 0.05, with protein-protein interaction significant at p < 1.1e-16 (Table S5) (Benjamini et al., 2001). One term, actin filament binding (GO:0051015) was enriched by a factor of 4 with FDR = 0.000000033. Of the 13 most significant GO terms (FDR < 0.0001), other notable enrichments included microfilament motor activity (GO:0000146) by a factor of 7, extracellular matrix structural constituent (GO:0005201) by a factor of 4, actin binding (GO:0003779) and ATP-dependent activity (GO:0140657), both by a factor of 2. An additional 11 GO terms had FDR < 0.001, of which cytoskeletal motor activity (GO:0003774) was the most prominent by a factor of 4. Fifteen GO terms were identified with FDR < 0.01 including spectrin binding (GO:0030507) by a factor of 8, myosin complex (GO:0016459) by a factor of 5, actin cytoskeleton (GO:0015629), actin filament-based process (GO:0030029), actin cytoskeleton organization (GO:0030036), and ion channel activity (GO:0005216), all by a factor of 2. Thirty-nine terms were enriched with FDR <0.05. Most remarkable among these included intracellular ligand-gated ion channel activity (GO:0005217) and axonemal dynein complex (GO:0005858) both by a factor of 6, stereocilium (GO:0032420), and stereocilium bundle (GO:0032421) both by a factor of 4. See Table S5 for a complete list of significantly enriched 78 GO terms in this set of 998 genes.
3.4 Variant level analysis
3.4.1 Variants found in affected infants only
We performed a comparative analysis between variants in our cohort and variants in genes found in other relevant studies (Gray et al., 2019; Halvorsen et al., 2021; Koh et al., 2022; Tester, Wong, Chanana, Jaye, et al., 2018) as well as OMIM terms associated with SIDS, Death in Infancy, Sudden Death, and SUDEP (Hamosh et al., 2022).
We report here 158 variants of interest in 93 infants (64.6% of the cohort) in 88 genes, including 130 rare/ultra-rare variants in 73 genes that have been previously associated with SIDS/SUID/SUDP. The majority of these variants, 117, were missense variants. Twelve variants occurred in the splice region as well as five splice acceptor and two splice donor variants. Ten variants resulted in a premature stop and nine resulted in a frameshift, while two induced in-frame insertions and one, an in-frame deletion. The number of variants per affected infant ranged from zero to six, with a mean number of 0.9 variants per infant. Three variants were recurrent. Two variants in CALM2 occurred in three infants, one was homozygous for one of these CALM2 variants, one infant was heterozygous for one CALM2 variant, and one infant was identified with both CALM2 variants. Additionally, two infants were found with the same CFTR variant. For individual genes, the number of variants ranged from 1 to 13. Fifty-eight infants were found with only one of these variants of interest, however, multiple variants were identified in several decedents. Twenty infants were found with two variants. Five infants were found with three variants, and four infants were found with four variants. Four infants had five of these variants, and two were found with six. Of the 88 genes in which variants of interest were found, 43 can be characterized as cardiac genes, 22 as neurologic, while 13 are related to systemic function, and two are associated with immunological function. An additional eight genes are related to various syndromes and are found in the Reactive Oxygen Species (ROS) pathway.
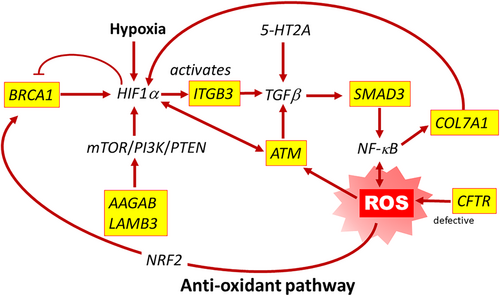
3.4.2 ROS pathway related genes (Table 2)
| Gene | Variant information | Allele frequency (AF) in gnomAD | Number of cases | Molecular consequence | Previously associated condition (ClinVar) |
|---|---|---|---|---|---|
| AAGAB | Associated OMIM conditions for the AAGAB variant below: Palmoplantar keratoderma punctate type 1A, AD, OMIM:148600 |
||||
| NC_000015.9:g.67528398G>A NM_024666.5(AAGAB):c.370C>T (p.Arg124*) | 0.00003120 (NE), 0.00002509 (Total AF) |
1 | Stop gained | Palmoplantar keratoderma punctate type 1A, P | |
| ATM | Associated OMIM conditions for all ATM variants below: Hereditary cancer predisposing syndrome, AD, OMIM:114480 Ataxia-telangiectasia syndrome, AR, OMIM:208900 |
||||
| NC_000011.9:g.108143447A>G | Not found (NE, Total AF) | 1 | Splice acceptor | Hereditary cancer predisposing syndrome, P; Ataxia-telangiectasia syndrome, P | |
NC_000011.9:g.108100002C>A NM_000051.4(ATM):c.283C>A (p.Gln95Lys) |
Not found (A) 0.0001240 (Total AF) |
1 | Missense | Hereditary cancer predisposing syndrome, LB/VUS; Ataxia-telangiectasia syndrome, LB/VUS |
|
| NC_000011.9:g.108172382G>C NM_000051.4(ATM):c.5185G>C (p.Val1729Leu) | 0.00004006 (A) 0.0001026 (Total AF) |
1 | Missense | Hereditary cancer predisposing syndrome, LB/VUS; Ataxia-telangiectasia syndrome, LB/VUS | |
| NC_000011.9:g.108178678T>A NM_001351834.2(ATM):c.5729T>A (p.Leu1910His) | Not found (NE, Total AF) | 1 | Missense | Not previously reported for this substitution Leu1910Pro at this location: Ataxia-telangiectasia syndrome, VUS | |
| NC_000011.9:g.108198447G>A NM_000051.4:c.7051G>A (p.Glu2351Lys) | Not found (A, Total AF) | 1 | Missense | Hereditary cancer predisposing syndrome, VUS | |
| NC_000011.9:g.108199766A>G NM_000051.4(ATM):c.7108A>G (p.Asn2370Asp) | Not found (A) 0.000007957 (Total AF) |
1 | Missense | Hereditary cancer predisposing syndrome, VUS; Ataxia-telangiectasia syndrome, VUS | |
| BRCA1 | Associated OMIM conditions for both BRCA1 variants below: Breast-ovarian cancer, familial, susceptibility to, AD, OMIM:604370 Fanconi anemia, AR, OMIM:617883 Pancreatic cancer, susceptibility to, 4, AD, OMIM:614320 |
||||
| NC_000017.10:g.41244546del NM_007294.4(BRCA1):c.3005del (p.Asn1002fs) | Not Found (A) 0.000003987 (Total AF) |
1 | Frameshift | Hereditary cancer-predisposing syndrome, P | |
| NC_000017.10:g.41246584C>G NM_007294.4(BRCA1):c.964G>C (p.Ala322Pro) | 0.00001759 (NE) 0.000007956 (Total AF) |
1 | Missense | Hereditary cancer-predisposing syndrome, LB/VUS; Breast-ovarian cancer, familial, susceptibility to, LB/VUS | |
| CFTR | Associated OMIM conditions for all CFTR variants below: Cystic fibrosis, AR, OMIM:219700 Bronchiectasis with or without elevated sweat chloride 1, modifier of, AD, OMIM:211400 Congenital bilateral absence of vas deferens, AR, OMIM:277180 Pancreatitis, chronic, susceptibility to, AD, OMIM:167800 |
||||
| NC_000007.13:g.117232712G>A NM_000492.4(CFTR):c.2490+1G>A | Not Found (A) 0.00002149 (NE) 0.00001050 (Total AF) |
2 | Splice donor | Cystic fibrosis, P; CFTR-related disorders, P | |
| NC_000007.13:g.117170992del NM_000492.4(CFTR):c.313del (p.Ile105fs) | 0.00006153 (A) 0.000003982 (Total AF) |
1 | Frameshift | Cystic fibrosis, P; Congenital bilateral aplasia of vas deferens from CFTR mutation, P; CFTR-related disorders, P | |
| NC_000007.13:g.117149200A>G NM_000492.4(CFTR):c.273+4A>G | 0.0002405 (A) 0.00002129 (Total AF) |
1 | Splice region | Cystic fibrosis, VUS; CFTR-related disorders, P/VUS | |
| NC_000007.13:g.117188691dup NM_000492.4(CFTR):c.451C>A (p.Gln151Lys) | Not found (A) 0.000007205 (Total AF) |
1 | Splice region | Cystic fibrosis, VUS; CFTR-related disorders, VUS | |
| COL7A1 | Associated OMIM Conditions for all COL7A1 variants below: Epidermolysis bullosa dystrophica/inversa AR, OMIM:226600 Toenail dystrophy, isolated, AD, OMIM:607523 Epidermolysis bullosa dystrophica, Bart type, AD, OMIM:132000 Transient bullous of the newborn, AD/AR, OMIM:131705 Epidermolysis bullosa dystrophica, AD, OMIM:131750 Epidermolysis bullosa pruriginosa, AD/AR, OMIM:604129 Epidermolysis bullosa, pretibial, OMIM:131850 |
||||
| NC_000003.11:g.48610345G>A NM_000094.4(COL7A1):c.6781C>T (p.Arg2261Ter) | 0.00000882 (NE) 0.000007968 (Total AF) |
1 | Stop gained | Epidermolysis bullosa dystrophica/inversa, P | |
| NC_000003.11:g.48611166G>A | Not found (NE, Total AF) | 1 | Splice region | Not previously reported | |
| NC_000003.11:g.48615751C>T | 0.000007763 (NE) 0.00002479 (Total AF) |
1 | Splice region | Not previously reported | |
| NC_000003.11:g.48629363C>T NM_000094.4(COL7A1):c.1325G>A (p.Arg442His) | Not found (AI/other) 0.0001556 (Total AF) |
1 | Missense | Not provided, VUS | |
| ITGB3 | Associated conditions in OMIM for the ITGB3 variant below: Glanzmann thrombasthenia 1, AD, OMIM:273800 Platelet-type bleeding disorder 16, AD, OMIM:187800 Myocardial infarction, susceptibility to, Inheritance mode not given, OMIM:608446 |
||||
| NC_000017.11:g.45384950:C:T NM_000212.3(ITGB3):c.2248C>T (p.Arg750Ter) | 0.0006008 (A) 0.00005657 (Total AF) |
1 | Stop gained | Glanzmann thrombasthenia, 1 and 2, P; Platelet-type bleeding disorder 16, P | |
| LAMB3 | Associated conditions in OMIM for the LAMB3 variant below: Amelogenesis imperfecta, type IA, AD, OMIM:104530 Epidermolysis bullosa junctional Herlitz type, AR, OMIM:226700 Epidermolysis bullosa junctional non-Herlitz type, AR, OMIM:226650 |
||||
| NC_000001.10:g.209801424G>A NM_000228.3(LAMB3):c.1244C>T (p.Pro415Leu) | 0.00002843 (NE) 0.00003406 (Total AF) |
1 | Missense | Not previously reported | |
| SMAD3 | Associated conditions in OMIM for the SMAD3 variant below: Loeys–Dietz syndrome 3, AD, OMIM:613795 |
||||
| NC_000015.9:g.67457303C>T NM_005902.4(SMAD3):c.277C>T (p.Arg93Ter) | 0.00001758 (NE) 0.000007955 (Total AF) |
1 | Stop gained | Familial thoracic aortic aneurysm and aortic dissection, P; Not provided, LP | |
- Abbreviations: A, African/African American; AD, autosomal dominant; AF, allele frequency; AI, American Indian; AR, autosomal recessive; B, benign; Hisp, Hispanic; LB, likely benign; LP, likely pathogenic; NE, Northern European; P, pathogenic; VUS, variant of uncertain significance.
The previously reported pathogenic variants included 20 in eight genes found in the antioxidant pathway that plays a critical role in the hypoxic response, the ROS pathway, AAGAB, ATM, BRCA1, CFTR, COL7A1, ITGB3, LAMB3, and SMAD3. (Figure 7). ROS pathway variants were identified in 20 infants (13.9% of the cohort). One infant had two different ROS pathway variants, one in ATM and one in CFTR. Two different infants were found with a duplicate CFTR variant, as previously stated. Exceptions to the inclusion of ultra-rare variants include two CFTR variants, and one ITBG3 variant. These variants were found in infants of African (A) descent and are more common in that population, not meeting our criteria for rare/ultra-rare. However, these variants are reported here due to the overall rarity of the allele in the combined total population and the previously reported pathogenicity in ClinVar. See Table 2. See Table S6 for details regarding the concurrence of the variants in each infant, corresponding demographic data and relevant ME notes.
3.4.3 Genes previously implicated in SUID
In addition to the ROS pathway variants, we detected 130 variants of interest in genes that were previously implicated in SIDS/SUID/SUDP (Baruteau et al., 2017; Chahal et al., 2020; Chahal et al., 2021; Clemens et al., 2020; Gray et al., 2019; Halvorsen et al., 2021; Keywan et al., 2021; Koh et al., 2022; Männikkö et al., 2018; Moore et al., 2020; Opdal, 2018; Rochtus et al., 2020; Tester, Wong, Chanana, Gray, et al., 2018; Tester, Wong, Chanana, Jaye, et al., 2018; Table 3). Eighty-eight of these variants occurred in 43 genes related to cardiac conditions including generalized cardiomyopathy, specific cardiomyopathies such as arrhythmogenic right ventricular dysplasia and left ventricular noncompaction; cardiac arrhythmias including atrial and ventricular fibrillation; the channelopathies, Brugada syndrome, Long QT syndrome and Catecholaminergic Polymorphic Ventricular Tachycardia (CPVT), in addition to several syndromes with a cardiac phenotype. Twenty-nine occurred in 22 genes related to neurologic function, including epilepsy, encephalopathy, ataxia, neuropathy and episodic pain syndrome, movement disorders such as dystonia and paralysis, intellectual disability and the polymalformation condition, Liang–Wang syndrome. Eleven additional variants were found in six genes related to systemic conditions and syndromes previously described in SIDS/SUID (Gray et al., 2019; Koh et al., 2022). Two variants were found in two genes related to immune function that have been previously studied in SIDS (Opdal, 2018). See Table 3.
| Gene | Variant information | Allele frequency (AF) in gnomAD | Number of cases | Molecular consequence | Previously associated condition (ClinVar) |
|---|---|---|---|---|---|
| Cardiac genes | |||||
| ABCC9 | Associated OMIM conditions for the ABCC9 variant below: Hypertrichotic osteochondrodysplasia, AD, OMIM:239850 Atrial fibrillation, familial 12, AD, OMIM:614050 Cardiomyopathy dilated, 10, AD, ClinVar (VUS), OMIM:608569 |
||||
NC_000012.11:g.21981893G>A NM_020297.4:(ABBC9)c.3668C>T (p.Thr1223Met) |
0.0000465 (NE) 0.00003537 (Total AF) |
1 | Missense | Not Specified, VUS; Cardiomyopathy dilated, 10, VUS | |
| ACTN2 | Associated OMIM conditions for the ACTN2 variant below: Myopathy, distal, 6, adult onset, AD, OMIM:618655 Cardiomyopathy, dilated, 1AA, with or without LVNC, AD, OMIM:612158 Cardiomyopathy, hypertrophic, 23, with or without LVNC, AD, OMIM:612158 Congenital myopathy 8, AD, OMIM:618654 |
||||
NC_000001.10:g.236883420A>G NM_001103.4(ACTN2):c.377A>G (p.Asn126Ser) |
Not found (A, Total AF) | 1 | Missense | Not previously reported | |
| AKAP9 | Associated OMIM conditions for all AKAP9 variants below: Long QT Syndrome, AD, OMIM:611820 |
||||
NC_000007.13:g.91630931C>T NM_005751.5(AKAP9):c.1700C>T (p.Thr567Ile) |
0.00002647 (NE) 0.00001197 (Total AF) |
1 | Missense | Long QT Syndrome, VUS | |
NC_000007.13:g.91699479C>G NM_005751.5(AKAP9):c.6466C>G (p.Leu2156Val) |
Not found (NE, Total AF) | 1 | Missense | Not previously reported | |
NC_000007.13:g.91732118dup NM_005751.5(AKAP9):c.11308dup (p.Arg3770LysfsTer28) |
Not found (A, Total AF) | 1 | Frameshift | Not previously reported | |
| AKAP10 | Associated OMIM conditions for the AKAP10 variant below: Cardiac Conduction Defect, AD, OMIM:115080 |
||||
NC_000017.10:g.19844264G>A NM_007202.4(AKAP10):c.1121C>T (p.Thr374Ile) |
Not found (A, Total AF) | 1 | Missense | Not previously reported | |
| ANK2 | Associated OMIM conditions for all ANK2 variants below: Cardiac arrhythmia, ankyrin-B-related, AD, OMIM: 600919 Long QT Syndrome 4, AD, OMIM: 600919 |
||||
NC_000004.11:g.114199044G>A NM_001148.6(ANK2):c.1735G>A (p.Ala579Thr) |
0.000008807 (NE) 0.00000398 (Total AF) |
1 | Missense | Not previously reported | |
NC_000004.11:g.114267060G>A NM_001148.6(ANK2):c.4253G>A (p.Arg1418His) |
0.00001762 (NE) 0.000007963 (Total AF) |
1 | Missense | Not previously reported | |
NC_000004.11:g.114280149C>T NM_001148.6(ANK2):c.10375C>T (p.Pro3459Ser) |
0.00002337 (NE) 0.00001067 (Total AF) |
1 | Missense | Long QT Syndrome, VUS; Cardiovascular phenotype, VUS | |
NC_000004.11:g.114294304A>G NM_001148.6(ANK2):c.11669A>G (p.Glu3890Gly) |
Not found (NE) 0.000003981 (Total AF) |
1 | Missense | Not previously reported | |
| BAG3 | Associated OMIM conditions for both BAG3 variants below: Cardiomyopathy, dilated, 1HH, AD, OMIM:613881 Myopathy, myofibrillar, 6, AD, OMIM:612954 |
||||
NC_000010.10:g.121411282C>T NM_004281.4(BAG3):c.95C>T (p.Pro32Leu) |
Not found (A, Total AF) | 1 | Missense | Not provided, VUS; Dilated cardiomyopathy 1HH, VUS; Myofibrillar myopathy 6, VUS | |
NC_000010.10:g.121436354G>A NM_004281.4(BAG3):c.1288G>A (p.Glu430Lys) |
Not found (NE, Total AF) | 1 | Missense | Myofibrillar myopathy 6, VUS; Dilated cardiomyopathy 1HH, VUS; Cardiovascular phenotype, VUS | |
| CACNA1C | Associated OMIM conditions for both CACNA1C variants below: Timothy Syndrome, AD, OMIM:601005 Long QT Syndrome, AD, OMIM:618447 Brugada Syndrome, AD, OMIM:611875 |
||||
NC_000012.11:g.2702441C>G NM_000719.7(CACNA1C):c.2593C>G (p.Leu865Val) |
Not found (NE) 0.000004023 (Total AF) |
1 | Missense | Long QT Syndrome, VUS | |
NC_000012.11:g.2800148G>A NM_000719.7(CACNA1C):c.6200G>A (p.Cys2067Tyr) |
Not found (NE) 0.000004472 (Total AF) |
1 | Missense | Not previously reported | |
| CACNB2 | Associated OMIM conditions for both CACNB2 variants below: Brugada Syndrome 4, AD, OMIM:611876 |
||||
| NC_000010.10:g.18439811G>A | Not found (NE, Total AF) | 1 | Splice acceptor | Not previously reported | |
NC_000010.10:g.18828300G>A NM_000724.4(CACNB2):c.1465G>A (p.Gly489Arg) |
Not found (NE, Total AF) | 1 | Missense | Not previously reported | |
| CALM2 | Associated OMIM conditions for both CALM2 variants below: Long QT syndrome 15, AD, OMIM:616249 |
||||
| NC_000002.11:g.47394863_47394867del | Not found (NE, Total AF) | 2 (Cmp Het, Het) | Splice Region | Not previously reported | |
| NC_000002.11:g.47394853_47394867del | Not found (A, NE, Total AF) | 2 (Cmp Het, Hom) | Splice Region | Not previously reported | |
| CTNNA3 | Associated OMIM conditions for the CTNNA3 variant below: Arrhythmogenic right ventricular dysplasia, familial 13, AD, OMIM:615616; |
||||
NC_000010.10:g.67680324C>T NM_013266.4(CTNNA3):c.2452G>A (p.Val818Ile) |
Not found (A, Total AF) | 1 | Missense | Not previously reported | |
| DCHS1 | Associated OMIM conditions for both DCHS1 variants below: Mitral valve prolapse 2, AD, OMIM:607829 Van Maldergem syndrome 1, AR, OMIM:601390 |
||||
NC_000011.9:g.6661424C>T NM_003737.4(DCHS1):c.1421G>A (p.Arg474His) |
0.00003123 (NE) 0.0000178 (Total AF) |
1 | Missense | Not previously reported | |
NC_000011.9:g.6661730G>A NM_003737.4(DCHS1):c.1115C>T (p.Ser372Phe) |
Not found (NE, Total AF) | 1 | Missense | Not previously reported | |
| DPP6 | Associated OMIM conditions for the DPP6 variant below: Intellectual Developmental Disorder, AD, OMIM:616311 Ventricular fibrillation, paroxysmal familial 2, AD, OMIM:612956 |
||||
NC_000007.13:g.154672617T>C NM_001936.5(DPP6):c.1912T>C (p.Tyr638His) |
Not found (A, Total AF) | 1 | Missense | Not previously reported | |
| DSG2 | Associated OMIM conditions for the DSG2 variant below: Arrhythmogenic right ventricular dysplasia 10, AD, OMIM:610193 Cardiomyopathy dilated 1BB, AR, OMIM:612877 |
||||
NC_000018.9:g.29104714A>T NM_001943.5(DSG2):c.877A>T (p.Ile293Leu) |
Not found (A) 0.00001068 (Total AF) |
1 | Missense | Arrhythmogenic right ventricular dysplasia 10, VUS; Cardiovascular Phenotype, VUS; Cardiomyopathy, VUS; Not specified, LB/VUS | |
| DSP | Associated OMIM conditions for all DSP variants below: Keratosis palmoplantaris striata II, AD, OMIM:612908 Arrhythmogenic right ventricular dysplasia 8, AD, OMIM:607450 Cardiomyopathy dilated with woolly hair and keratoderma, AR, OMIM:605676 Skin fragility-woolly hair syndrome, AR, OMIM:607655 Dilated cardiomyopathy with woolly hair keratoderma and tooth agenesis, AD, OMIM:615821 Epidermolysis bullosa lethal acantholytic, AR, OMIM:609638 |
||||
NC_000006.11:g.7558461G>A NM_004415.4(DSP):c.386G>A (p.Arg129Gln) |
0.00001759 (NE) 0.00001193 (Total AF) |
1 | Missense | Cardiomyopathy, VUS; Not provided, VUS | |
NC_000006.11:g.7572209G>A NM_004415.4:c.2038G>A (p.Glu680Lys) |
Not found (NE, Total AF) | 1 | Missense | Arrhythmogenic cardiomyopathy with woolly hair and keratoderma, VUS; Arrhythmogenic right ventricular dysplasia 8, VUS; Cardiovascular phenotype, VUS | |
NC_000006.11:g.7577256T>C NM_004415.4(DSP):c.2858T>C (p.Leu953Pro) |
0.00002654 (NE) 0.000026544 (Total AF) |
1 | Missense | Cardiomyopathy, VUS; Cardiovascular Phenotype, VUS; Not provided, VUS | |
NC_000006.11:g.7583856G>A NM_004415.4(DSP):c.6361G>A (p.Gly2121Arg) |
8.796E-06 (NE) 0.000003978 (Total AF) |
1 | Missense | Arrhythmogenic cardiomyopathy with woolly hair and keratoderma, VUS; Arrhythmogenic right ventricular dysplasia 8, VUS | |
NC_000006.11:g.7585410C>T NM_004415.4(DSP):c.7915C>T (p.Arg2639Trp) |
0.00003898 (Unk, Total AF) | 1 | Missense | Cardiomyopathy, VUS; Arrhythmogenic cardiomyopathy with woolly hair and keratoderma, VUS; Arrhythmogenic right ventricular dysplasia 8, VUS; Not provided, VUS | |
| FLNC | Associated OMIM conditions for the FLNC variant below: Cardiomyopathy, familial hypertrophic 26, AD, OMIM:617047 Myopathy myofibrillar 5, AD, OMIM:609524 Cardiomyopathy, familial restrictive 5, AD, OMIM:617047 Myopathy, distal 4, AD, OMIM:614065 |
||||
NC_000007.13:g.128482877C>T NM_001458.5(FLNC):c.2419C>T (p.Pro807Ser) |
0.00002659 (NE) 0.00001205 (Total AF) |
1 | Missense | Not provided, VUS; Cardiovascular phenotype, VUS; Inborn genetic diseases, VUS; Dilated Cardiomyopathy, Dominant, VUS; Hypertrophic cardiomyopathy 26, VUS; Distal myopathy with posterior leg and anterior hand involvement, VUS; Myofibrillar myopathy 5, VUS | |
| FPGT-TNNI3 | Associated OMIM conditions for the FPGT-TNNI3 variant below: Cardiac conduction disease with or without dilated cardiomyopathy, AD, OMIM:616117 |
||||
NC_000001.10:g.75009665G>A NM_015978.3(TNNI3K):c.2507G>A (p.Ter836=extTer7) |
Not found (Hisp, Total AF) | 1 | Stop gained | Not provided, VUS | |
| GJA1 | Associated OMIM conditions for both GJA1 variants below: Erythrokeratodermia variabilis et progressiva 3, AD, OMIM:617525 Craniometaphyseal dysplasia, AR, OMIM:218400 Atrioventricular septal defect 3, AD, OMIM:600309 Oculodentodigital dysplasia, AD, OMIM:164200; AR, OMIM:257850 Syndactyly type III, AD, OMIM: 186100 Hypoplastic left heart syndrome 1, AR, OMIM: 241550 Palmoplantar keratoderma with congenital alopecia, AD, OMIM:104100 |
||||
NC_000006.11:g.121768451G>A NM_000165.5(GJA1):c.458G>A (p.Arg153Gln) |
Not found (A, Total AF) | 1 | Missense | Not previously reported | |
NC_000006.11:g.121768843C>T NM_000165.5(GJA1):c.850C>T (p.Pro284Ser) |
Not found (NE, Total AF) | 1 | Missense | Not previously reported | |
| HCN4 | Associated OMIM conditions for the HCN4 variant below: Brugada syndrome 8, AD, OMIM:613123 Sick sinus syndrome 2, AD, OMIM:163800 |
||||
NC_000015.9:g.73615240A>G NM_005477.3(HCN4):c.3194T>C (p.Val1065Ala) |
Not found (A, Total AF) | 1 | Missense | Brugada syndrome 8, VUS | |
| JPH2 | Associated OMIM conditions for the JPH2 variant below: Cardiomyopathy hypertrophic 17, AD, OMIM:613873 |
||||
NC_000020.10:g.42788880G>C NM_020433.5(JPH2):c.547C>G (p.Pro183Ala) |
Not found (A, Total AF) | 1 | Missense | Hypertrophic cardiomyopathy, VUS; Hypertrophic cardiomyopathy 17, VUS; Cardiomyopathy, Dilated, 2E, VUS; Cardiovascular phenotype, VUS | |
| KCND2 | No Associated OMIM conditions for the KCND2 variant below. | ||||
NC_000007.13:g.120373105C>T NM_012281.3(KCND2):c.1264C>T (p.Arg422*) |
Not found (NE, Total AF) | 1 | Stop gained | Not previously reported | |
| KCNH2 | Associated OMIM conditions for all KCNH2 variants below: Long QT syndrome 2, acquired susceptibility to, AD, OMIM:613688 Long QT syndrome 2, AD, OMIM:613688 Short QT syndrome 1, mode of inheritance not given, OMIM:609620 |
||||
NC_000007.13:g.150655492_150655500dup NM_000238.4(KCNH2):c.567_575dup (p.Gly192_Ala193insAlaProGly) |
0.000003987 (Unk, Total AF) | 1 | In-frame insertion | Long QT syndrome, VUS; Cardiovascular phenotype, VUS; Not provided, VUS | |
| LAMA4 | Associated OMIM conditions for all LAMA4 variants below: Cardiomyopathy, dilated 1JJ, AD, OMIM:615235 |
||||
NC_000006.11:g.112441568C>T NM_002290.5(LAMA4):c.4562G>A (p.Arg1521His) |
0.00002647 (NE) 0.00004784 (Total AF) |
1 | Missense | Cardiomyopathy, dilated 1JJ, VUS; Cardiovascular phenotype, VUS; Not provided, VUS | |
NC_000006.11:g.112441622T>C NM_002290.5(LAMA4):c.4508A>G (p.Tyr1503Cys) |
Not found (NE, total AF) | 1 | Missense | Not previously reported | |
NC_000006.11:g.112454633A>T NM_002290.5(LAMA4):c.3593T>A (p.Phe1198Tyr) |
Not found (NE, total AF) | 1 | Missense | Not previously reported | |
| LDB3 | Associated OMIM conditions for both LDB3 variants below: Cardiomyopathy hypertrophic 24, AD, OMIM:601493 Cardiomyopathy dilated 1C with or without LVNC, AD, OMIM:601493 Myopathy, myofibrillar 4, AD, OMIM:609452 Left ventricular noncompaction 3, AD, OMIM:601493 |
||||
NC_000010.10:g.88446961G>A NM_001080114.2(LDB3):c.480G>A (p.Met160Ile) |
Not found (NE, Total AF) | 1 | Missense | Myofibrillar myopathy 4, VUS | |
NC_000010.10:G:Ag.88459055G>A NM_001080115.2(LDB3):c.917G>A (p.Arg306His) |
0.00001563 (NE) 0.00001428 (Total AF) |
1 | Missense | Myofibrillar myopathy 4, VUS | |
| MYBPC3 | Associated OMIM conditions for both MYBPC3 variants below: Cardiomyopathy hypertrophic, AR/AD, OMIM:115197 Cardiomyopathy dilated 1MM, AD, OMIM:615396 Left ventricular noncompaction 10, AD, OMIM:615396 |
||||
NC_000011.9:g.47353764C>T NM_000256.3(MYBPC3):c.3673G>A (p.Ala1225Thr) |
Not found (NE) 0.000004013 (Total AF) |
1 | Missense | Not provided, VUS; Cardiomyopathy, VUS; Hypertrophic cardiomyopathy, VUS | |
NC_000011.9:g.47365086C>T NM_000256.3(MYBPC3):c.1180G>A (p.Val394Ile) |
0.000009127 (NE) 0.000004138 (Total AF) |
1 | Missense | Hypertrophic cardiomyopathy, VUS | |
| MYH6 | Associated OMIM conditions for both MYH6 variants below: Atrial septal defect 3, Inheritance mode not given, OMIM:614089 Cardiomyopathy, hypertrophic 14, AD, OMIM:613251 Sick sinus syndrome 3, mode of inheritance not given, OMIM:614090 Cardiomyopathy dilated 1EE, AD, OMIM:613252 |
||||
NC_000014.8:g.23852520C>T NM_002471.4(MYH6):c.5575G>A (p.Asp1859Asn) |
0.00001644 (NE) 0.000007666 (Total AF) |
1 | Missense | Hypertrophic cardiomyopathy 14, VUS; Cardiovascular Phenotype, VUS; Not provided, VUS | |
NC_000014.8:g.23855760C>T NM_002471.4(MYH6):c.4723G>A (p.Glu1575Lys) |
0.000007745 (NE) 0.00002476 (Total AF) |
1 | Missense | Hypertrophic cardiomyopathy 14, VUS; Cardiovascular Phenotype, VUS | |
| MYH11 | Associated OMIM conditions for all MYH11 variants below: Aortic aneurysm, familial thoracic 4, AD, OMIM:132900 |
||||
NC_000016.9:g.15797934G>A NM_002474.3(MYH11):c.5833C>T (p.Arg1945Cys) |
0.00000879 (NE) 0.000007953 (Total AF) |
1 | Missense | Familial thoracic aortic aneurysm and aortic dissection, VUS; Not provided, VUS | |
NC_000016.9:g.15815461T>G NM_002474.3(MYH11):c.4396A>C (p.Lys1466Gln) |
0.000008791 (NE) 0.000003977 (Total AF) |
1 | Missense | Aortic aneurysm, familial thoracic 4, VUS; Familial thoracic aortic aneurysm and aortic dissection, VUS | |
| NC_000016.9:g.15847245C>T | Not Found (NE, Total AF) | 1 | Splice Region | Familial thoracic aortic aneurysm and aortic dissection, LB | |
| NC_000016.9:g.15850194G>C | Not Found (NE) 0.0000354 (Total AF) |
1 | Splice Region | Aortic aneurysm, familial thoracic 4, VUS; Cardiovascular phenotype, VUS; Familial thoracic aortic aneurysm and aortic dissection, VUS | |
NC_000016.9:g.15857704C>T NM_002474.3(MYH11):c.1078G>A (p.Val360Ile) |
0.00001551 (NE) 0.00005308 (Total AF) |
1 | Missense | Aortic aneurysm, familial thoracic 4, VUS; Familial thoracic aortic aneurysm and aortic dissection, LB/VUS; Inborn genetic diseases, VUS | |
| MYLK | Associated OMIM conditions for both MYLK variants below: Aortic aneurysm, familial thoracic 7, AD, OMIM:613780 Megacystis-microcolon-intestinal hypoperistalsis syndrome, AR, OMIM:249210 |
||||
| NC_000003.11:g.123444927T>C | 0.00002663 (NE) 0.000012 (Total AF) |
1 | Splice Acceptor | Not provided, VUS/LP | |
NC_000003.11:g.123457763G>A NM_053025.4(MYLK):c.569C>T (p.Pro190Leu) |
0.00002365 (NE) 0.00006481 (Total AF) |
1 | Missense | Aortic aneurysm, familial thoracic 7, VUS; Familial thoracic aortic aneurysm and aortic dissection, VUS | |
| MYPN | Associated OMIM conditions for the MYPN variant below: Cardiomyopathy, dilated 1KK, AD, OMIM:615248 Cardiomyopathy familial restrictive, 4, AD, OMIM:615248 Nemaline myopathy 11, AR, OMIM:617336 Cardiomyopathy, hypertrophic 22, AD, OMIM: 615248 |
||||
NC_000010.10:g.69881260C>G NM_032578.4(MYPN):c.65C>G (p.Ala22Gly) |
Not found (A) 0.0002546 (Total AF) |
1 | Missense | Dilated cardiomyopathy 1KK, LB; Cardiovascular Phenotype, LB; Not provided, VUS | |
| NOTCH1 | Associated OMIM conditions for both NOTCH1 variants below: Aortic valve disease 1, AD, OMIM:109730 Adams–Oliver syndrome 5, AD, OMIM: 616028 |
||||
NC_000009.11:g.139400291C>T NM_017617.5(NOTCH1):c.4057G>A (p.Gly1353Ser) |
0.00004519 (A) 0.00001583 (Total AF) |
1 | Missense | Not provided, VUS | |
NC_000009.11:g.139417452C>T NM_017617.5(NOTCH1):c.592G>A (p.Glu198Lys) |
Not found (NE) 0.00001023 (Total AF) |
1 | Missense | Not previously reported | |
| PRDM16 | Associated OMIM conditions for the PRDM16 variant below: Left ventricular noncompaction 8, AD, OMIM:615373 Cardiomyopathy, dilated, 1LL, AD, OMIM:615373 |
||||
NC_000001.10:g.3347438C>T NM_022114.4(PRDM16):c.3287C>T (p.Ala1096Val) |
Not found (A) 0.00002836 (Total AF) |
1 | Missense | Left ventricular noncompaction 8, VUS; Not provided, VUS | |
| RANGRF; SLC25A35 | No Associated OMIM conditions for the RANGRF/SLC25A35 variant below. | ||||
NC_000017.10:g.8192285C>G NM_016492.5(RANGRF):c.89C>G (p.Pro30Arg) |
Not found (A) 0.000003984 (Total AF) |
1 | Missense | Not previously reported | |
| RYR2 | Associated OMIM conditions for all RYR2 variants below: Ventricular tachycardia catecholaminergic polymorphic 1, AD, OMIM:604772 Arrhythmogenic right ventricular dysplasia 2, AD, OMIM:600996 |
||||
NC_000001.10:g.237496932A>T NM_001035.3(RYR2):c.2440C>A (p.Leu814Ile) |
Not found (NE, Total AF) | 1 | Splice region | Not previously reported | |
NC_000001.10:g.237666632C>A NM_001035.3(RYR2):c.2440C>A (p.Leu814Ile) |
Not found (A, Total AF) | 1 | Missense | Not previously reported | |
NC_000001.10:g.237670022C>A NM_001035.3(RYR2):c.2626C>A (p.Pro876Thr) |
Not found (NE, Total AF) | 1 | Missense | Not previously reported | |
NC_000001.10:g.237870348G>A NM_001035.3(RYR2):c.9680G>A (p.Arg3227His) |
Not found (A) 0.00000713 (Total AF) |
1 | Missense | Catecholaminergic polymorphic ventricular tachycardia, VUS; Cardiovascular phenotype, VUS | |
NC_000001.10:g.237921044A>G NM_001035.3(RYR2):c.11293A>G (p.Ile3765Val) |
0.00004132 (A) 0.000007134 (Total AF) |
1 | Missense | Catecholaminergic polymorphic ventricular tachycardia, VUS; Cardiovascular phenotype, VUS | |
NC_000001.10:g.237957284G>A NM_001035.3(RYR2):c.13900G>A (p.Val4634Ile) |
0.0000143 (Unk, Total AF) | 1 | Missense | Arrhythmogenic right ventricular dysplasia 2, VUS | |
| SCN1B | Associated OMIM conditions for the SCN1B variant below: Atrial fibrillation familial 13, AD, OMIM:615377 Developmental and epileptic encephalopathy 52, AR, OMIM:617350 Cardiac conduction defect nonspecific generalized with febrile seizures plus type 1, AD, OMIM:604233 Brugada syndrome 5, AD, OMIM:612838 |
||||
NC_000019.9:g.35524842G>A NM_199037.5(SCN1B):c.647G>A (p.Gly216Asp) |
Not found (A, Total AF) | 1 | Missense | Brugada syndrome 5, VUS; Not specified, VUS | |
| SCN3B | Associated OMIM conditions for the SCN3B variant below: Brugada syndrome 7, AD, OMIM:613120 Atrial fibrillation, familial 16, AD, OMIM:613120 |
||||
NC_000011.9:g.123513271C>T NM_018400.4(SCN3B):c.328G>A (p.Val110Ile) |
0.00003871 (NE) 0.0003111 (Total AF) |
1 | Missense | Death in Infancy, P; Cardiomyopathy, LB; Brugada syndrome 7, LB; Cardiovascular phenotype, LB; Not provided, LB, VUS | |
| SCN5A | Associated OMIM conditions for the SCN5A variant below: Atrial fibrillation, familial 10, AD, OMIM:614022 Sick sinus syndrome 1, AD, OMIM: 608567 Sudden infant death syndrome, susceptibility to, AR, OMIM:272120 Ventricular fibrillation, familial 1, mode of inheritance not given, OMIM:603829 Long QT syndrome 3, AD, OMIM:603830 Heart block, nonprogressive, AD, OMIM:113900 Cardiomyopathy, dilated, 1E, AD, OMIM:601154 Brugada syndrome 1, AD, OMIM:601144 Heart block, progressive, type IA, AD, OMIM:113900 |
||||
NC_000003.11:g.38591816A>T NM_000335.5(SCN5A):c.6044T>A (p.Val2015Glu) |
Not found (NE, Total AF) | 1 | Missense | Not previously reported | |
| SCN10A | Associated OMIM conditions for both SCN10A variants below: Episodic pain syndrome, familial 2; OMIM:615551 |
||||
NC_000003.11:g.38770056C>T NM_006514.4(SCN10A):c.2617G>A (p.Val873Met) |
0.00002761 (NE) 0.00001246 (Total AF) |
1 | Missense | Brugada Syndrome, VUS; Not provided, VUS | |
NC_000003.11:g.38770187C>T NM_006514.4(SCN10A):c.2486G>A (p.Arg829His) |
0.000008803 (NE) 0.000007959 (Total AF) |
1 | Missense | Brugada Syndrome, VUS | |
| SLMAP | No Associated OMIM conditions for the SLMAP variant below. | ||||
NC_000003.11:g.57882621A>G NM_007159.5(SLMAP):c.1361A>G (p.Asp454Gly) |
0.000008848 (NE) 0.000004015 (Total AF) |
1 | Missense | Not previously reported | |
| TGFB3 | Associated OMIM conditions for the TGFB3 variant below: Loeys–Dietz syndrome 5, AD, OMIM:615582 Arrhythmogenic right ventricular dysplasia 1, AD, OMIM:107970 |
||||
NC_000014.8:g.76431926G>A NM_003239.5(TGFB3):c.754+5C>T |
Not found (NE, Total AF) | 1 | Splice Region | Rienhoff syndrome, VUS; Not provided, LB; Familial thoracic aortic aneurysm and aortic dissection, VUS | |
| TMEM43 | Associated OMIM conditions for the TMEM43 variant below: Emery-Dreifuss muscular dystrophy 7, AD, OMIM:614302 Arrhythmogenic right ventricular dysplasia 5, AD, OMIM:604400 |
||||
NC_000003.11:g.14173126T>C NM_024334.3(TMEM43):c.344T>C (p.Leu115Pro) |
0.0000264 (NE) 0.00001194 (Total AF) |
1 | Missense | Cardiovascular phenotype, VUS; Not provided, VUS; Cardiomyopathy, VUS; Arrhythmogenic right ventricular dysplasia 5, VUS | |
| TNNC1 | Associated OMIM conditions for the TNNC1 variant below: Cardiomyopathy hypertrophic 13, AD, OMIM:13243 Cardiomyopathy dilated 1Z, AD, OMIM:611879 |
||||
NC_000003.11:g.52486116G>A NM_003280.3(TNNC1):c.202+6C>T |
Not found (A) 0.000003984 (Total AF) |
1 | Splice Region | Dilated cardiomyopathy 1Z, VUS; Hypertrophic cardiomyopathy 13, VUS | |
| TRDN | Associated OMIM conditions for both TRDN variants below: Ventricular tachycardia catecholaminergic polymorphic 5 with or without muscle weakness, AR, OMIM:615441 |
||||
NC_000006.11:g.123869697dup NM_006073.4(TRDN):c.295dup |
Not found (A, Total AF) | 1 | Missense | Not previously reported | |
NC_000006.11:g.123869698C>T NM_006073.4(TRDN):c.292G>A (p.Glu98Lys) |
Not found (A, Total AF) | 1 | Missense | Cardiovascular Phenotype, VUS | |
| TRPM4 | Associated OMIM conditions for the TRPM4 variant below: Erythrokeratodermia veriabilis et progressiva 6, AD, OMIM:618531 Progressive familial heart block, type IB, AD, OMIM:604559 |
||||
NC_000019.9:g.49671868_49671869insGAA NM_017636.4(TRPM4):c.671_672insGAA (p.Pro224_Leu225insAsn) |
Not found (NE, Total AF) | 1 | Inframe insertion | Not previously reported | |
| TTN | Associated OMIM conditions for all TTN variants below: Myopathy myofibrillar 9 with early respiratory failure, AD, OMIM:603689 Cardiomyopathy familial hypertrophic 9, AD, OMIM:613765 Muscular dystrophy limb-girdle autosomal recessive 10, AR, 608807 Cardiomyopathy dilated, 1G, OMIM:604145 Tibial muscular dystrophy, tardive, AD, OMIM:600334 Salih myopathy, AR, OMIM:611705; |
||||
NC_000002.11:g.179393283T>C NM_003319.4(TTN):c.80000A>G (p.Glu26667Gly) |
Not found (NE, Total AF) | 1 | Missense | Not previously reported | |
NC_000002.11:g.179404843C>T NM_003319.4(TTN):c.70855G>A (p.Gly23619Ser) |
Not found (A, Total AF) | 1 | Missense | Not previously reported | |
NC_000002.11:g.179418773G>A NM_003319.4(TTN):c.61870C>T (p.Arg20624Cys) |
Not found (A) 0.000008046 (Total AF) |
1 | Missense | Not provided, VUS | |
NC_000002.11:g.179419446A>G NM_003319.4(TTN):c.61433T>C (p.Phe20478Ser) |
Not found (A, Total AF) | 1 | Missense | Not previously reported | |
NC_000002.11:g.179422192C>T NM_003319.4(TTN):c.60602G>A (p.Gly20201Asp) |
Not found (Unk, Total AF) | 1 | Missense | Not previously reported | |
NC_000002.11:g.179422617A>G NM_003319.4(TTN):c.60269T>C (p.Val20090Ala) |
Not found (NE, Total AF) | 1 | Missense | Not previously reported | |
NC_000002.11:g.179440747C>T NM_003319.4(TTN):c.42917G>A (p.Arg14306His) |
Not found (NE) 0.00000805 (Total AF) |
1 | Missense | Not previously reported | |
NC_000002.11:g.179463580G>T NM_003319.4(TTN):c.29662C>A (p.Leu9888Ile) |
Not found (A, Total AF) | 1 | Missense | Not previously reported | |
NC_000002.11:g.179479442C>G NM_003319.4(TTN):c.21604G>C (p.Gly7202Arg) |
Not found (A, Total AF) | 1 | Missense | Not previously reported | |
NC_000002.11:g.179604015T>C NM_003319.4(TTN):c.12856A>G (p.Lys4286Glu) |
Not found (NE, Total AF) | 1 | Missense | Dilated cardiomyopathy 1G, VUS; Autosomal recessive limb-girdle muscular dystrophy type 2J, VUS | |
NC_000002.11:g.179613353C>A NM_133379.5(TTN):c.13774G>T (p.Glu4592*) |
0.000008910 (NE) 0.000004007 (Total AF) |
1 | Stop gained | Not previously reported | |
NC_000002.11:g.179636193C>T NM_003319.4(TTN):c.7723G>A (p.Ala2575Thr) |
Not found (A, Total AF) | 1 | Missense | Not previously reported | |
NC_000002.11:g.179640399A>C NM_003319.4(TTN):c.6054T>G (p.Phe2018Leu) |
0.00003553 (NE) 0.00001599 (Total AF) |
1 | Missense | Dilated cardiomyopathy 1G, VUS; Autosomal recessive limb-girdle muscular dystrophy type 2, VUS; Not provided, VUS | |
| Neurological genes | |||||
| CACNA1H | Associated OMIM conditions for all CACNA1H variants below: Epilepsy, idiopathic generalized, susceptibility to, 6, OMIM:611942 Epilepsy, childhood absence, susceptibility to, 6, OMIM:611942 Hyperaldosteronism, familial, type IV, AD, OMIM:617027 Epilepsy, idiopathic generalized, susceptibility to, 1, AD, OMIM:600669 |
||||
NC_000016.9:g.1203949C>G NM_021098.3(CACNA1H):c.212C>G (p.Pro71Arg) |
Not found (NE, Total AF) | 1 | Missense | Idiopathic generalized epilepsy, VUS; Hyperaldosteronism, familial, type IV, VUS; Epilepsy, childhood absence, susceptibility to, 6, VUS | |
NC_000016.9:g.1250576G>T NM_021098.3(CACNA1H):c.1119+5G>T |
Not found (A, Total AF) | 1 | Splice region | Not previously reported | |
NC_000016.9:g.1264987C>G NM_021098.3(CACNA1H):c.4945C>G (p.Leu1649Val) |
Not found (A, Total AF) | 1 | Missense | Not previously reported | |
NC_000016.9:g.1270539_1270548del NM_021098.3(CACNA1H):c.6607_6616del (p.Ser2203ProfsTer11) |
Not found (NE, Total AF) | 1 | Frameshift | Not previously reported | |
| CACNB4 | Associated OMIM conditions for the CACNB4 variant below: Epilepsy, idiopathic generalized, susceptibility to, 9, AD, OMIM:607682; Episodic ataxia, type 5, AD, OMIM:613855; |
||||
NC_000002.11:g.152695823G>T NM_000726.5(CACNB4):c.1373C>A (p.Ser458Tyr) |
Not found (A) 0.00002010 (Total AF) |
1 | Missense | Idiopathic generalized epilepsy, VUS | |
| CHD2 | Associated OMIM conditions for the CHD2 variant below: Developmental and epileptic encephalopathy 94, AD, OMIM:602119 |
||||
NC_000015.9:g.93567817C>A NM_001271.4(CHD2):c.5369C>A (p.Pro1790His) |
Not found (A) 0.00001603 (Total AF) |
1 | Missense | Not previously reported | |
| CLCN4 | Associated OMIM conditions for the CLCN4 variants below: Raynaud-Claes syndrome, X-linked dominant, OMIM:300114 |
||||
NC_000023.10:g.10181730C>T NM_001830.4(CLCN4):c.1586C>T (p.Thr529Ile) |
Not found (A, Total AF) | 1 | Missense | Not previously reported | |
| DNM1L | Associated OMIM conditions for the DNM1L variant below: Encephalopathy, lethal due to defective mitochondrial peroxisomal fission 1, AR, AD, OMIM:614388 Optic atrophy 5, AD, OMIM: 610708; |
||||
NC_000012.11:g.32871615A>C NM_005690.5(DNM1L):c.658A>C (p.Met220Leu) |
Not found (A, Total AF) | 1 | Missense | Inborn genetic diseases, VUS | |
| DYRK1A | Associated OMIM conditions for the DYRK1A variant below: Intellectual Developmental Disorder, AD, OMIM:614104 |
||||
NC_000021.8:g.38850482G>C NM_001347721.2(DYRK1A):c.208-28G>C |
0.0001231 (A) 0.000007971 (Total AF) |
1 | Splice Acceptor | DYRK1A-related intellectual disability syndrome, VUS; Not provided, VUS | |
| EFHC1 | Associated OMIM conditions with the EFHC1 variant below: Myoclonic epilepsy juvenile susceptibility to 1, AD, OMIM:254770 Epilepsy juvenile absence susceptibility to, AD, OMIM:607631 |
||||
NC_000006.11:g.52303214T>C NM_018100.4(EFHC1):c.398T>C (p.Val133Ala) |
Not found (A, Total AF) | 1 | Missense | Not previously reported | |
| GRIN1 | Associated OMIM conditions with the GRIN1 variant below: Urodevelopmental disorder with or without hyperkinetic movements and seizures, AR, OMIM:617820 Neurodevelopmental disorder with or without hyperkinetic movements and seizures, AD, OMIM:614254 |
||||
NC_000009.11:g.140056463A>G NM_000832.7(GRIN1):c.1555A>G (p.Ile519Val) |
Not found (A, Total AF) | 1 | Missense | Not previously reported | |
| GRIN2A | Associated OMIM conditions with the GRIN2A variant below: Epilepsy focal with speech disorder and with or without impaired intellectual development, AD, OMIM:245570 |
||||
NC_000016.9:g.9858026C>G NM_000833.5(GRIN2A):c.212C>G (p.Ala71Gly) |
Not found (A, Total AF) | 1 | Missense | Landau–Kleffner syndrome, VUS Not provided, VUS | |
| GRIN2B | Associated OMIM conditions with the GRIN2B variant below: Developmental and epileptic encephalopathy 27, AD, OMIM:616139 Intellectual Developmental Disorder, autosomal dominant 6, AD, OMIM:613970 |
||||
NC_000012.11:g.13761764G>C NM_000834.5(GRIN2B):c.1783C>G (p.Pro595Ala) |
Not found (A) 0.000007967 (Total AF) |
1 | Missense | Not previously reported | |
| KCNA2 | Associated OMIM conditions for the KCNMA1 variant below: Developmental and epileptic encephalopathy 32, AD, OMIM:616366 |
||||
NC_000001.10:g.111144730dup NM_001204269.2(KCNA2):c.957dup (p.Asp320*) |
Not found (NE, Total AF) | 1 | Frameshift | Not previously reported | |
| KCNMA1 | Associated OMIM conditions related to both KCNMA1 variants below: Liang–Wang syndrome, AD, OMIM:618729 Epilepsy idiopathic generalized susceptibility to 16, AD, OMIM:618596 Cerebellar atrophy and seizures, AR, OMIM:617643 Paroxysmal nonkinesigenic dyskinesia 3 with or without generalized epilepsy, AD, OMIM:609446 |
||||
NC_000010.10:g.78734333T>G NM_002247.4(KCNMA1):c.2093-4508A>C |
Not found (A) 0.000006762 (Total AF) |
1 | Splice acceptor | Not previously reported | |
NC_000010.10:g.78674702G>A NM_002247.4(KCNMA1):c.2834C>T (p.Thr945Ile) |
Not found (Unk, Total AF) | 1 | Missense | Not previously reported | |
| KCNQ3 | Associated OMIM conditions for the KCNQ3 variant below: Seizures benign neonatal 2, AD, OMIM:121201 |
||||
NC_000008.10:g.133146633A>G NM_004519.4(KCNQ3):c.1703T>C (p.Ile568Thr) |
Not found (A, Total AF) | 1 | Missense | Not previously reported | |
| KCNT1 | Associated OMIM conditions for the KCNT1 variant below: Epilepsy nocturnal frontal lobe 5, AD, OMIM:615005 Developmental and epileptic encephalopathy 14, AD, OMIM:614959 |
||||
NC_000009.11:g.138671256G>A NM_020822.3(KCNT1):c.2781G>A (p.Met927Ile) |
Not found (NE, Total AF) | 1 | Missense | Not previously reported | |
| RANBP2 | Associated OMIM conditions for the RANBP2 variant below: Encephalopathy acute infection-induced 3 susceptibility to, AD, OMIM:608033 |
||||
NC_000002.11:g.109382226G>A NM_006267.5(RANBP2):c.5231G>A (p.Cys1744Tyr) |
Not found (NE, Total AF) | 1 | Missense | Not previously reported | |
| SCN1A | Associated OMIM conditions for both SCN1A variants below: Febrile Seizures, Familial, 3A, AD, OMIM:604403 Migraine, familial hemiplegic, 3, AD, OMIM:609634 Dravet syndrome, AD, OMIM:607208 Epilepsy, generalized, with febrile seizures plus, Type 2, AD, OMIM:604403 |
||||
NC_000002.11:g.166848857A>G NM_006920.6(SCN1A):c.4895T>C (p.Ile1632Thr) |
Not found (A, Total AF) | 1 | Missense | Not previously reported | |
NC_000002.11:g.166892993G>T NM_006920.6(SCN1A):c.2961C>A (p.Asp987Glu) |
Not found (NE, Total AF) | 1 | Missense | Severe myoclonic epilepsy in infancy, LP; Not provided, VUS | |
| SCN2A | Associated OMIM conditions for the SCN2A variant below: Episodic ataxia type 9, AD, OMIM:618924 Developmental and epileptic encephalopathy 11, AD, OMIM:613721 Seizures benign familial infantile 3, AD, OMIM:607745 |
||||
NC_000002.11:g.166246035G>A NM_021007.3(SCN2A):c.5719G>A (p.Val1907Met) |
Not found (NE) 0.000003989 (Total AF) |
1 | Missense | Not previously reported | |
| SCN4A | Associated OMIM conditions for the SCN4A variant below: Hyperkalemic periodic paralysis type 2, AD, OMIM:170500 Paramyotonia congenita, AD, OMIM:168300 Myotonia congenita atypical acetazolamide-responsive, AD, OMIM:608390 Myasthenic syndrome congenital 16, AR, OMIM:614198 Hypokalemic periodic paralysis type 2, AD, OMIM:613345 |
||||
NC_000017.10:g.62022878A>G NM_000334.4(SCN4A):c.3562T>C (p.Tyr1188His) |
Not found (A) 0.000003988 (Total AF) |
1 | Missense | Inborn genetic diseases, VUS; Not provided, VUS; Hyperkalemic periodic paralysis, VUS | |
| SCN8A | Associated OMIM conditions for the SCN8A variant below: Developmental and epileptic encephalopathy 13, AD, OMIM:614558 Cognitive impairment with or without cerebellar ataxia, AD, OMIM:614306 Myoclonus, familial 2, AD, OMIM:618364 |
||||
NC_000012.11:g.52115618A>T NM_014191.4(SCN8A):c.1924A>T (p.Thr642Ser) |
Not found (A, Total AF) | 1 | Missense | Not previously reported | |
| SCN9A | Associated OMIM conditions for both SCN9A variants below: Neuropathy hereditary sensory and autonomic type IID, AR, OMIM:243000 Small fiber neuropathy, AD, OMIM:133020 Paroxysmal extreme pain disorder, OMIM:167400 Insensitivity to pain, congenital, AR, OMIM:243000 Erythermalgia primary, AD, OMIM:133020 |
||||
NC_000002.11:g.167055652C>T NM_002977.3(SCN9A):c.5464G>A (p.Gly183Ser) |
0.00001758 (NE) 0.000007953 (Total AF) |
1 | Missense | Not provided, VUS; Inborn genetic diseases, VUS; Neuropathy, hereditary sensory and autonomic, type 2A, VUS; Generalized epilepsy with febrile seizures plus, type 7, VUS | |
NC_000002.11:g.167128959T>C NM_002977.3(SCN9A):c.3268A>G (p.Met1090Val) |
Not found (A) 0.00001224 (Total AF) |
1 | Missense | Not provided, VUS Inborn genetic diseases, VUS; Neuropathy, hereditary sensory and autonomic, type 2A, VUS Generalized epilepsy with febrile seizures plus, type 7, VUS | |
| SCN11A | Associated OMIM conditions for the SCN11A variants below: Neuropathy hereditary sensory and autonomic type VII, OMIM:615548 Episodic pain syndrome, familial 3, OMIM:615552 |
||||
NC_000003.11:g.38941513C>T NM_014139.3(SCN11A):c.1894G>A (p.Asp632Asn) |
Not found (NE, Total AF) | 1 | Missense | Not previously reported | |
NC_000003.11:g.38924807G>A NM_014139.3(SCN11A):c.3136C>T (p.Arg1046Trp) |
Not found (A) 0.000007961 (Total AF) |
1 | Missense | Not previously reported | |
| SLC2A1 | Associated OMIM conditions for the SLC2A1 variant below: Dystonia 9, AD, OMIM:601042 GLUT1 deficiency syndrome 1 infantile onset severe, AR, OMIM:606777 Stomatin-deficient cryohydrocytosis with neurologic defects, AD, OMIM:608885 GLUT1 deficiency syndrome 2, childhood onset, AD, OMIM:612126 Epilepsy, idiopathic generalized susceptibility to, AD, OMIM:614847 |
||||
NC_000001.10:g.43392783C>T NM_006516.4(SLC2A1):c.1408G>A (p.Gly470Arg) |
Not found (A) 0.000003983 (Total AF) |
1 | Missense | Not provided, VUS; GLUT1 deficiency syndrome 1, autosomal recessive, VUS | |
| Immunological/inflammatory genes | |||||
| IL6 | Associated OMIM conditions with the IL6 variant below: Intracranial hemorrhage in brain cerebrovascular malformations susceptibility to, SM, OMIM:108010 Rheumatoid arthritis systemic juvenile, NG, OMIM:604302 Type 1 diabetes mellitus, AR, OMIM:222100 Type 2 diabetes mellitus, AD, OMIM:125853 Kaposi sarcoma susceptibility to, AD, OMIM:148000 Crohn disease-associated growth failure, MU, OMIM:266600 |
||||
NC_000007.13:g.22768315T>C NM_000600.5(IL6):c.214T>C (p.Cys72Arg) |
0.00006163 (A) 0.000003982 (Total AF) |
1 | Missense | Not previously reported | |
| IL1RN | Associated OMIM conditions with the IL1RN variant below: Gastric cancer risk after H. pylori infection, AD, OMIM:137215 Microvascular complications of diabetes 4, NG, OMIM:612628 Chronic recurrent multifocal osteomyelitis 2, with periostitis and pustulosis, AR, OMIM:612852 Interleukin 1 receptor antagonist deficiency, AR, OMIM:147679 |
||||
NC_000002.11:g.113890332G>A NM_000577.5(IL1RN):c.364G>A (p.Ala122Thr) |
0.00002324 (NE) 0.0001698 (Total AF) |
1 | Missense | Sterile multifocal osteomyelitis with periostitis and pustulosis, VUS | |
| Systemic/syndromic genes | |||||
| CHRNA1 | Associated OMIM conditions with the CHRNA1 variant below: Myasthenic syndrome, congenital, 1A, slow-channel, AD, OMIM:601462 Myasthenic syndrome, congenital, 1B, fast-channel, AD, OMIM:608930 Multiple pterygium syndrome, lethal type, AR, OMIM:253290 |
||||
NC_000002.11:g.175619130G>C NM_000079.4(CHRNA1):c.357C>G (p.Asp119Glu) |
Not found (NE) 0.000003985 (Total AF) |
1 | Missense | Not provided, VUS; Lethal multiple pterygium syndrome, VUS | |
| FLNA | Associated OMIM conditions with both FLNA variants below: Otopalatodigital syndrome type I, X-linked dominant; OMIM:311300 Congenital short bowel syndrome, X-linked recessive, OMIM:300048 Otopalatodigital syndrome type II, OMIM:304120 Intestinal pseudoobstruction, neuronal, X-linked recessive; OMIM:300048 Melnick–Needles syndrome, X-linked dominant, OMIM:309350 Cardiac valvular dysplasia, X-linked, OMIM:314400 FG syndrome 2, X-linked, OMIM:300321 Heterotopia, Periventricular 1, X-linked dominant OMIM:300049 Terminal osseous dysplasia, X-linked dominant, OMIM:300244 Frontometaphyseal dysplasia 1, X-linked recessive, OMIM:305620 |
||||
NC_000023.10:g.153588821G>C NM_001456.4(FLNA):c.3342C>G (p.Cys1114Trp) |
Not found (A, Total AF) | 1 | Missense | Oto-palato-digital syndrome, type II, VUS; Frontometaphysea dysplasia, VUS; Heterotopia, periventricular, X-linked dominant, VUS; Melnick–Needles syndrome, VUS | |
NC_000023.10:g.153577231C>T NM_001456.4(FLNA):c.7906G>A (p.Val2636Ile) |
0.00001285 (NE) 0.000005687 (Total AF) |
1 | Missense | Developmental delay, VUS | |
| PHOX2B | Associated OMIM conditions with the PHOX2B variant below: Neuroblastoma with Hirschsprung disease, OMIM:613013 Central hypoventilation syndrome congenital with or without Hirschsprung disease, AD, OMIM:209880 |
||||
NC_000004.11:g.41750401C>G NM_003924.4(PHOX2B):c.227G>C (p.Ser76Thr) |
0.0001976 (Hisp) 0.00003907 (Total AF) |
1 | Missense | Hereditary cancer-predisposing syndrome, B; Not provided, VUS; Congenital central hypoventilation, VUS; Neuroblastoma, susceptibility to, 2, VUS; Haddad syndrome, VUS | |
| POLG | Associated OMIM conditions with the POLG variant below: Progressive external ophthalmoplegia with DNA deletions, AD, OMIM: 157640 Mitochondrial DNA depletion syndrome 4B (MNGIE type), AR, OMIM:613662 Mitochondrial recessive ataxia syndrome (includes SANDO and SCAE), AR, OMIM:607459 Mitochondrial DNA depletion syndrome 4A (Alpers type), AR, OMIM:203700 Progressive external ophthalmoplegia 1, AR, OMIM:258450 |
||||
NC_000015.9:g.89862458C>T NM_002693.3(POLG):c.3104+1G>A |
0.0001202 (A) 0.00001061 (Total AF) |
1 | Splice donor | Progressive sclerosing poliodystrophy, LP/P Not provided, LP | |
| TBX5 | Associated OMIM conditions with both TBX5 variant below: Holt–Oram syndrome, AD, OMIM:142900 |
||||
NC_000012.11:g.114841675A>C NM_000192.3(TBX5):c.29T>G (p.Leu10Arg) |
Not found (Hisp, Total AF) | 1 | Missense | Not previously reported | |
NC_000012.11:g.114793873G>C NM_000192.3(TBX5):c.1021C>G (p.Pro341Ala) |
Not found (A, Total AF) | 1 | Missense | Cardiovascular phenotype, VUS | |
| TSC1 | Associated OMIM conditions with all TSC1 variants below: Tuberous sclerosis-1, AD, OMIM:191100 Focal cortical dysplasia type II somatic, OMIM:607341 Lymphangioleiomyomatosis, OMIM:606690 |
||||
NC_000009.11:g.135772112T>A NM_000368.5(TSC1):c.3005A>T (p.Asp1002Val) |
0.00002677 (NE) 0.00006454 (Total AF) |
1 | Missense | Hereditary cancer-predisposing syndrome, LB; Not provided, LB; Tuberous sclerosis 1, VUS; Isolated focal cortical dysplasia type II, LB | |
NC_000009.11:g.135772578C>T NM_000368.5(TSC1):c.2968G>A (p.Glu990Lys) |
0.00002639 (NE) 0.00006365 (Total AF) |
1 | Missense | Hereditary cancer-predisposing syndrome, LB; Not provided, LB; Tuberous sclerosis 1, VUS; Isolated focal cortical dysplasia type II, LB | |
NC_000009.11:g.135781212G>A NM_000368.5(TSC1):c.1753C>T (p.Pro585Ser) |
Not found (NE) 0.000003982 (Total AF) |
1 | Missense | Hereditary cancer-predisposing syndrome, VUS; Tuberous sclerosis LB; 1, Not provided, VUS | |
NC_000009.11:g.135786914_135786916del NM_000368.5(TSC1):c.957_959del (p.Leu320del) |
Not found (NE, Total AF) | 1 | Inframe deletion | Tuberous sclerosis 1, LB; Not provided, LB/VUS; Seizure, VUS; Hereditary cancer-predisposing syndrome, VUS | |
- Abbreviations: A, African/African American; AD, autosomal dominant; AF, allele frequency; AI, American Indian; AR, autosomal recessive; B, benign; Hisp, Hispanic; LB, likely benign; LP, likely pathogenic; MU, multifactorial inheritance; NE, Northern European; NG, inheritance not given; P, pathogenic; SM, somatic mutation; VUS, variant of uncertain significance.
Cardiac: Cardiomyopathy
Thirty-nine variants in 17 genes associated with cardiomyopathies and related conditions were found in 35 infants—five infants had two of these variants. These include ABCC9, ACTN2, BAG3, DSG2, DSP, FLNC, JPH2, LAMA4, LDB3, MYBPC3, MYH6, MYPN, PRDM16, SCN5A, TNNC1, FPGT-TNNI3K, and TTN. Five variants in three of these genes, JPH2, LAMA4, and TNNC1, were found in five infants and are associated with various cardiomyopathies in the absence of other conditions (OMIM:613873, OMIM:615235, OMIM:613243, and OMIM:611879). One LAMA4 variant, Arg1521His, has been previously reported in ClinVar as a variant of unknown significance (VUS) for cardiomyopathy and an unspecified cardiovascular phenotype (ClinVar:213607). The infant that was found with this variant had a family history of SIDS. Eighteen variants in five genes associated with cardiomyopathy and myopathy, ACTN2, BAG3, FLNC, MYPN, and TTN, were found in 18 infants (OMIM:613881, OMIM:612954, OMIM:617047, OMIM:614065, OMIM:617336, OMIM:615248, OMIM:618655, OMIM:612158, OMIM:618654). One of the infants with one of the TTN variants was noted to have cardiac complications at birth that were expected to resolve with age. Five variants in three genes, LDB3, MYBPC3, and PRDM16, associated with cardiomyopathy and left ventricular noncompaction were identified in five infants (OMIM:601493, OMIM:609452, OMIM:115197, OMIM:615396, OMIM:615373). Six variants in two genes, DSG2 and DSP were identified in six different children that are associated with cardiomyopathy and right ventricular dysplasia (OMIM:610193, OMIM:612877, OMIM:607450, OMIM:605676). Two variants in two infants were found in ABCC9 and SCN5A that are associated with both cardiomyopathy and atrial fibrillation (OMIM:608569, OMIM:614050, OMIM:601154, OMIM:614022). The variant in SCN5A is also associated with ventricular fibrillation (OMIM:603829), Brugada (OMIM:601144), Long QT syndrome (OMIM:603830), heart blocks (OMIM:113900), Sick Sinus syndrome (OMIM:608567), and susceptibility to SIDS (OMIM:272120). One variant in one gene associated with cardiomyopathy and a cardiac conduction disorder, FPGT-TNNI3K, was found in one infant (OMIM:616117). Two MYH6 variants were found in two different infants that were related to cardiomyopathy (OMIM:613251, OMIM:613252), Sick Sinus syndrome (OMIM:614090), and cardiac defects including atrial septal defect (OMIM:614089). One infant in our cohort was found to have atrial septal defect at autopsy and another had an unspecified heart defect, but these variants did not occur in either of these infants. There were no autopsy reports available for the infants that carried these MYH6 variants.
Cardiac: Congenital conditions
Thirteen variants in five genes, DCHS1, GJA1, MYH11, MYLK, and NOTCH1, were found in 11 infants related to heart defects and congenital conditions in the absence of cardiomyopathy. Six variants were found in five infants in three genes associated with heart defects, DCHS1, GJA1, and NOTCH1. These included mitral valve prolapse (OMIM:607829), atrioventricular septal defect (OMIM:606215), and aortic valve disease (OMIM:109730). One infant had a variant in both GJA1 and NOTCH1. An autopsy report was available for only one of these five infants and no heart defect was found. Five MYH11 variants associated with familial thoracic aortic aneurysm were found in four infants, one infant had two of these MYH11 variants (OMIM:132900). Two MYLK variants, were found in two infants and are associated with familial thoracic aortic aneurysm (OMIM:613780). One missense (Pro190Leu) variant is reported to be a VUS for aortic aneurysm in ClinVar (ClinVar:520017). The child with the Pro190Leu variant had a family history of SIDS. There were no relevant findings reported at autopsy other than the presence of focal intimal thickening in the right coronary artery.
Cardiac: Arrhythmias and conduction disorders
Five variants were found in five genes associated with cardiac arrhythmias and conduction disorders in the absence of cardiomyopathy such as cardiac conduction defect, AKAP10 (OMIM:115080), arrhythmogenic right ventricular dysplasia, CTNNA3 (OMIM:615616), ventricular fibrillation, DPP6 (OMIM:612956), arrhythmogenic right ventricular dysplasia, TGFB3 (OMIM:107970), and TMEM43 (OMIM:604400). These variants were found in five infants.
Channelopathies: Brugada syndrome, Long QT syndrome, and Catecholaminergic Polymorphic Ventricular Tachycardia (CPVT)
Thirty-two variants were found in 27 decedents in 17 genes related to the channelopathies Brugada syndrome, Long QT syndrome, and CPVT. Nine variants in seven genes, CACNB2, HCN4, KCND2, RANGRF, SCN10A, SLMAP, and TRPM4 were identified in eight decedents that have been implicated in Brugada syndrome in previous studies (Baruteau et al., 2017; Keywan et al., 2021; Koh et al., 2022; Rochtus et al., 2020; Tester, Wong, Chanana, Jaye, et al., 2018 (OMIM:611876, OMIM:613123)). Two of these variants in genes related to Brugada syndrome, SCN10A and SLMAP, were found in one infant. The SCN10A variant has been previously reported in ClinVar as a VUS for Brugada syndrome (ClinVar:641909) and also associated with the neurologic condition, episodic pain syndrome (OMIM:615551). The SLMAP variant has not been previously reported. Three variants in SCN1B, SCN3B, and SCN5A found in three infants, have been previously associated with Brugada syndrome as well as other arrhythmias (OMIM:612838, OMIM:613120, OMIM:601144). The SCN3B variant found has also been previously reported as P for death in infancy (ClinVar:190886). The SCN5A variant as previously discussed in Section 3.4.3.1, Cardiac: Cardiomyopathy, has been associated with Long QT and SIDS among other conditions. The mother of the child in which the SCN1B variant was found was noted to have lost another child at three months of age to SIDS. Two infants were found with two additional variants in one gene, CACNA1C, that has been associated with Brugada syndrome (Chahal et al., 2020; Halvorsen et al., 2021; Koh et al., 2022; OMIM:611875) in addition to other arrhythmias (OMIM:601005, OMIM:609620), and Long QT syndrome (OMIM:618447). Six variants were found in three genes in seven infants that have previously been linked to Long and/or Short QT Syndrome alone, AKAP9, CALM2, and KCNH2 (Baruteau et al., 2017; Chahal et al., 2020; Clemens et al., 2020; Keywan et al., 2021; Koh et al., 2022; Moore et al., 2020; Tester, Wong, Chanana, Gray, et al., 2018; Tester, Wong, Chanana, Jaye, et al., 2018; OMIM:611820, OMIM:616249, OMIM:613688, OMIM:609620). Four variants in ANK2 were found in four infants. Variants in this gene have been linked to Long QT syndrome in addition to other arrhythmias previously (Baruteau et al., 2017; Chahal et al., 2020; Koh et al., 2022; OMIM:600919). Six variants in RYR2 were found in six infants. Variants in RYR2 have been associated with CPVT (Baruteau et al., 2017; Chahal et al., 2020; Chahal et al., 2021; Halvorsen et al., 2021; Koh et al., 2022; Tester, Wong, Chanana, Jaye, et al., 2018, OMIM:604772) and arrhythmogenic right ventricular dysplasia (OMIM:600996). Five decedents were found to have more than one of these variants related to arrhythmias. One infant was found with two variants, one in SCN10A and another in ANK2. Another had a variant in both CACNA1C and AKAP9, and a third infant had two different CALM2 variants as well as a variant in ANK2. A fourth was homozygous for one of the CALM2 variants and also had a variant in RYR2. Two variants in TRDN were identified in one infant. Variants in TRDN have been associated with CPVT through an AR mode of inheritance in OMIM (OMIM:615441).
3.4.4 Neurologic disorders
Twenty-nine variants were found in 22 genes in 26 infants previously implicated in SIDS/SUID/SUDP that are related to neurologic disorders. Three infants had two of these variants each. Eleven variants in eleven infants were found in seven genes related to epilepsy, four in CACNA1H, one each in CACNB4, EFHC1, GRIN1, GRIN2A, KCNQ3, and two in SCN1A. (Baruteau et al., 2017; Keywan et al., 2021; Koh et al., 2022; Rochtus et al., 2020; Tester, Wong, Chanana, Gray, et al., 2018; Tester, Wong, Chanana, Jaye, et al., 2018; OMIM:611942, OMIM:600669, OMIM:607682, OMIM:254770, OMIM:607631, OMIM:614254, OMIM:245570, OMIM:121201, OMIM:604403, and OMIM:607208). One SCN1A missense variant, Asp987Glu, has been previously reported in ClinVar to be LP for severe myoclonic epilepsy in infancy (SMEI), also known as Dravet syndrome (ClinVar:224116), a condition which has been previously implicated in SIDS/SUID/SUDP and SUDEP (Chahal et al., 2020; Chahal et al., 2021; Gray et al., 2019; Keywan et al., 2021; Koh et al., 2022; Rochtus et al., 2020). Five variants were found in four genes in five children related to epilepsy and other conditions KCNMA1, KCNT1, SCN2A, and SLC2A1 (OMIM:618596, OMIM:617643, OMIM:609446, OMIM:615005, OMIM:614959, OMIM:613721, OMIM:607745, OMIM:618924, OMIM:614847, OMIM:608885) Two infants were found with two different KCNMA1 variants associated with the polymalformation condition, Lian–Wang syndrome (OMIM:618729). The infant with the KCNQ3 variant also had a variant in SLC2A1 associated with dystonia (OMIM:601042), childhood onset of GLUT1 deficiency syndrome 2 of AD inheritance (OMIM:612126), Stomatin-deficient cryohydrocytosis with neurologic defects, AD, (OMIM:608885) as well as epilepsy (OMIM:614847). This infant was noted to have a “well developed infant brain with isolated microglial nodules” by the ME. Five variants in five genes were found in five infants related to encephalopathy, CHD2, DNM1L, GRIN2B, KCNA2, and RANBP2 (OMIM:615369, OMIM:614388, OMIM:616139, OMIM:616366, OMIM:608033). One SCN8A variant found in one infant was associated with both epilepsy and encephalopathy (OMIM:614558, OMIM:618364). The infant with the RANBP2 variant also had the KCNT1 variant, related to epilepsy (OMIM:615005), as previously stated, and encephalopathy (OMIM:614959). The infant with the DNM1L variant was noted at autopsy to have “neuronal changes consistent with hyperacute hypoxia ischemia.” Two additional infants were found to have one variant each in SCN11A, related to episodic pain syndrome and neuropathy (OMIM:615552, OMIM:615548). Another two children were each found with a variant in SCN9A also related to neuropathy (OMIM:243000, OMIM:133020). Two variants were identified in two children related to intellectual disability, CLCN4 and DYRK1A (OMIM:300114, OMIM:614104) The infant with the DYRK1A variant also had the KCNMA1 variant. Finally, one SCN4A variant was identified in one infant related to movement disorders and paralysis (OMIM:168300, OMIM:608390, OMIM:170500, OMIM:613345). This variant is reported as VUS in ClinVar for Hyperkalemic periodic paralysis and other conditions (ClinVar:1899177).
3.4.5 Systemic/syndromic conditions
Nineteen variants were found in thirteen genes in fourteen infants, most of which are reported to be VUS, LP, or P for various syndromes for which there was no noted diagnosis at the time of death. Variants in six of these genes have been previously implicated in SIDS/SUID/SUDP, including CHRNA1, FLNA, PHOX2B, POLG, TBX5, and TSC1 (Koh et al., 2022; Opdal & Rognum, 2011; Tables 3 and 4). The variant in CHRNA1 has been linked to Lethal multiple pterygium syndrome, (ClinVar:466178). The variant in FLNA has been associated with multiple conditions in OMIM including, Otopalatodigital syndrome (OMIM:311300), Melnick–Needles syndrome (OMIM:309350), Cardiac valvular dysplasia (OMIM:314400), FG syndrome (OMIM:300321), Periventricular heterotopia (OMIM:300049), Terminal osseous dysplasia (OMIM:300244). The variant in PHOX2B has been implicated in Central hypoventilation syndrome (VUS, ClinVar:486030, OMIM:209880). The POLG variant has been associated with Progressive sclerosing poliodystrophy (LP, ClinVar:619308) and Progressive external ophthalmoplegia with DNA deletions (OMIM:157640). The TBX5 variant has been previously associated with Holt–Oram syndrome (OMIM:142900). Both of the TSC1 variants have been associated with Tuberous Sclerosis (B/LB/VUS, ClinVar:237715, B/LB/VUS, ClinVar:237714, OMIM:191100), Focal cortical dysplasia (OMIM:607341) and Lymphangioleiomyomatosis (OMIM:606690). The infant in which the PHOX2B variant was identified was noted to be “in the 10th percentile of growth for age,” and to have “mesenteric lymphadenopathy, hepatosplenomegaly, adrenal hypoplasia” at autopsy. This infant was also found to have one of the TBX5 variants. The child in which the CHRNA1 variant was found was also found to have a DYNC1H1 variant (discussed in the next paragraph) previously reported as P in ClinVar for spinal muscular atrophy, a condition of AD inheritance (ClinVar:30034). This child was noted to have been hospitalized for 10 days following birth for unspecified reasons. See Tables 3 and S6.
| Gene | Variant information | Allele frequency (AF) in gnomAD | Number of cases | Molecular consequence | Previously reported related condition (ClinVar) |
|---|---|---|---|---|---|
| ALB | Associated OMIM Conditions for the ALB variant below: Dysalbuminemic hyperthyroxinemia, AD, OMIM:615999 Analbuminemia, AD, OMIM:616000 |
||||
| NC_000004.11:g. 74274452C>T NM_000477.7(ALB):c.412C>T (p.Arg138Ter) | 0.0000155 (NE) 0.000007075 (Total AF) |
1 | Stop gained | Congenital analbuminemia, P; Analbuminemia Bethesda, P | |
| C12orf65/ MTRFR | Associated OMIM Conditions for the C12orf65 variant below: Spastic paraplegia, combined oxidative phosphorylation deficiency 7, OMIM:613559 |
||||
| NC_000012.11:g.123738317_123738320dup NM_152269.5(MTRFR):c.96_99dup (p.Pro34fs) | 0.00008521 (NE) 0.00003890 (Total AF) |
1 | Frameshift | Combined oxidative phosphorylation defect type 7 Spastic paraplegia, P; Not Provided, P | |
| DYNC1H1 | Associated OMIM Conditions for the DYNC1H1 variant below: Cortical dysplasia, complex, with other brain malformations 13, AD, OMIM:614563 Spinal muscular atrophy lower extremity-predominant 1, AD, OMIM:158600 Charcot-Marie-Tooth disease, axonal, type 20, AD, OMIM:614228 |
||||
| NC_000014.8:g. 102457904A>G NM_001376.5(DYNC1H1):c.2909A>G (p.Tyr970Cys) | Not found (NE, Total AF) | 1 | Missense | Spinal muscular atrophy, lower extremity-predominant, 1, autosomal dominant, P | |
| GPR98/ ADGRV1 | Associated OMIM Conditions for both GPR98/ADGRV1 variants below: Febrile seizures, familial, 4, AD, OMIM:604352 Usher syndrome, type 2C, GPR98/PDZD7 digenic, AR, OMIM:605472 |
||||
| NC_000005.9:g.89986808C>T NM_032119.4(ADGRV1):c.6901C>T (p.Gln2301Ter) | 0.0001091 (NE) 0.00004993 (Total AF) |
1 | Stop gained | Febrile seizures, familial, 4 Usher syndrome type 2C, P; Usher syndrome type 2C, LP/P; Rare genetic deafness, P; Not provided, P | |
| NC_000005.9:g. 90119376C>A NM_032119.4(ADGRV1):c.16331C>A (p.Thr5444Lys) | 0.0006544 (NE) 0.0003458 (Total AF) |
1 | Missense | Febrile seizures, familial, 4 Usher syndrome type 2C, VUS; Not provided, VUS | |
| HBA2 | Associated OMIM Conditions for the HBA2 variant below: Thalassemia alpha, OMIM:604131 Erythrocytosis 7, AD, OMIM:617981; Heinz body anemia, AD, OMIM:140700 Hemoglobin H disease, deletional and nondeletional, OMIM:613978 |
||||
| NC_000016.9:g.223515del NM_000517.6(HBA2):c.345del (p.Ala116fs) | 0.0000447 (A) 0.000007227 (Total AF) |
1 | Frameshift | Not provided, P | |
| PIEZO2 | Associated OMIM Conditions for PIEZO2 variant below: Arthrogryposis distal with impaired proprioception and touch, AR, OMIM:617146; Arthrogryposis distal Type 3, AD, OMIM:114300; Arthrogryposis distal Type 5, AD, OMIM:108145 Marden–Walker syndrome, AD, OMIM:248700 |
||||
| NC_000018.9:g.10680240del NM_001378183.1(PIEZO2):c.7911del (p.Asn2638fs) | 0.0000176 (NE) 0.00001195 (Total AF) |
1 | Frameshift deletion | Inborn genetic diseases, P | |
| SPG7 | Associated OMIM Conditions for the SPG7 variant below: Spastic paraplegia 7, autosomal recessive, (AD/AR), OMIM:607259 |
||||
| NC_000016.9:g. 89576947T>A NM_003119.4(SPG7):c.233T>A (p.Leu78Ter) | 0.0001550 (NE) 0.0003964 (Total AF) |
1 | Stop gained | Spastic Parapalegia 7, P | |
- Abbreviations: A, African/African American; AD, autosomal dominant; AF, allele frequency; AI, American Indian; AR, autosomal recessive; B, benign; Hisp, Hispanic; LB, likely benign; LP, likely pathogenic; NE, Northern European; P, Pathogenic; VUS, variant of uncertain significance.
Eight of the nineteen variants involved in systemic function were incidentally found in seven genes in seven of these fourteen infants, that have not been reported in SIDS/SUID previously. These rare (<0.01%) variants are reported to be associated in ClinVar and OMIM with various syndromes, ALB (P, Congenital analbuminemia, ClinVar:156320, OMIM:616000), (Dysalbuminemic hyperthyroxinemia, OMIM:615999); C12orf65/MTHFR (P, Spastic paraplegia, ClinVar:214192, OMIM:613559); DYNC1H1 (P, Spinal muscular atrophy, ClinVar:30034, OMIM:158600), GPR98/ADGRV1 (P, Usher Syndrome, Febrile Seizures, ClinVar:6798, OMIM:604352, OMIM:605472); HBA2 (P, Not provided, ClinVar:439116, Thalassemia alpha, OMIM:604131, Hemoglobin H disease, OMIM:613978, Heinz body anemia, OMIM: 140700); PIEZO2 (P, Inborn genetic diseases, ClinVar:521668, Arthrogryposis distal, OMIM:114300, OMIM:108145, OMIM:617146, Marden-Walker syndrome, OMIM:248700); and SPG7 (Spastic Parapalegia, P, ClinVar:6816, OMIM:607259); Two GPR98/ADGRV1 variants in one infant were identified that are related to familial febrile seizures and Usher syndrome, both of digenic and AR inheritance. The first results in a premature stop and has been previously reported to be P for these conditions (ClinVar:6798). The second variant is a missense variant that is reported to be a VUS for these conditions (ClinVar:198666). This variant is found more commonly in the Northern European (NE) population (AF = 0.065%, NE; Total AF = 0.035%) and does not meet our criteria for rare. (The infant in which this variant was found was white.) Due to the mode of inheritance of these conditions, this variant is also likely significant for the development of disease, so both are included here. See Tables 4 and S6.
3.4.6 Immunological function
Two variants in two infants were found in two genes that have been previously examined in SIDS/SUID studies. The first, an IL-6 missense variant, has been associated with two conditions with AD inheritance, type 2 diabetes mellitus (OMIM:125853) and susceptibility to Kaposi sarcoma (OMIM:148000). This variant has also been associated with Crohn disease-associated growth failure in a multi-factorial (MU) pattern of inheritance (OMIM:266600), one AR inherited disorder, type 1 diabetes mellitus (OMIM:222100) and one condition, systemic juvenile rheumatoid arthritis for which no inheritance pattern is given (NG). In addition, somatic mutations (SM) in this allele have been linked to susceptibility to intracranial hemorrhage in brain cerebrovascular malformations (OMIM:108010). This infant was reported to have no findings at autopsy. Notably, this infant also had a variant in IL4R, but this variant did not pass our rigorous filtering process, though it has been associated with immune conditions inherited in an AD manner. It was found to be slightly too common in the overall population (0.058%) and much more common in populations of African descent (0.607%) to be included. The infant in which this variant was found was described as African American. One variant in IL-1RN (also known as IL-1Rα), which encodes the interleukin-1 receptor antagonist, was found in the second infant. This variant has been linked to gastric cancer risk after H. pylori infection (OMIM:137215) in an AD manner and also to microvascular complications of diabetes 4 (OMIM:612628, inheritance pattern NG) In addition, it has been associated with chronic recurrent multifocal osteomyelitis 2 with periostitis and pustulosis (OMIM:612852) and interleukin 1 receptor antagonist deficiency (OMIM:147679), both with an AR mode of inheritance. Genetic irregularities in IL-1Rα have been previously described in SIDS (Opdal, 2018).
3.4.7 Group analysis: Median age at death
Infants were grouped according to the type and number of variants each were found to have, cardiac, immunologic, neurologic, ROS pathway, and systemic/syndromic. The median age of death was calculated for each group among children that were found to have only one variant. A single variant in a cardiac related gene was found in 29 infants. Fourteen infants were found with a single variant related to neurologic function, and 11 infants were identified with a single ROS pathway gene variant. There were only two infants with one variant among the immunologic and systemic/syndromic groups, so these groups were excluded from the analysis. The median age at death for 29 infants with a single cardiac gene variant was 10.1 weeks (IQR = 8.9, 13.0), whereas for the 14 infants that were found to have a single variant in a gene related to neurologic function, the median age at death was 16.1 weeks (IQR = 11.1, 21.2). The 11 infants with a single variant in a ROS pathway gene died at the median age of 12.7 weeks (IQR = 9.5, 21.5). ANOVA was run in R and revealed that these differences were marginally significant (p = 0.053) (Figure 8). See Table S7 for a full list of abbreviations used in this manuscript.
4 DISCUSSION
We set out to explore the potential contribution of gene variants to the etiology of SUID. We identified numerous variants in genes associated with SUID, remarkably in 64.6% of our cohort. Some of these are variants known for their involvement in cardiac diseases, neurological disorders, the immune response, the regulation of neuronal excitability and the response to hypoxia and oxidative stress (ROS). Gene enrichment studies in affected infants further confirmed the validity of these findings.
4.1 ROS pathway variants
Each of the genes in the ROS pathway in which P/LP variants were identified contributes to the regulation of the response to hypoxia, in particular the activation of HIF1α and the production of ROS (Yuan et al., 2013). For example, AAGAB, highly expressed in the brain, regulates PTEN nuclear translocation to promote the functional neuronal recovery following hypoxic-ischemic induced brain injury (Dai et al., 2021). ATM is activated by hypoxia and re-oxygenation and thus acts as a redox sensor (Hammond & Giaccia, 2004). It plays a critical role in maintaining cellular redox homeostasis (Hammond & Giaccia, 2004). Dysfunction in ATM has been implicated in heart failure and sudden death. BRCA1 plays a major role in the hypoxic response by regulating HIF-1α stability, and also rescues neurons from cerebral ischemia/perfusion injury through the NRF2 signaling pathway (Xu et al., 2018). CFTR is well studied in cystic fibrosis—and associated with respiratory failure, but CFTR also functions as a regulator of ROS and has been associated with impairment in oxidative phosphorylation (Ntimbane et al., 2009). CFTR belongs to the ATP binding cassette (ABC) transporter superfamily and is also widely expressed in the CNS (Vasiliou et al., 2009). COL7A1 is also a regulator in the hypoxic response and HIF-1α, and it is activated by NF-kB (Amelio et al., 2018). ITGB3 is required for sustained TGF-β pathway activation (Sesé et al., 2017). It is activated by HIF1α in hypoxia and its knockdown has been associated with increased apoptosis and reduced neuronal survival and migration, particularly under hypoxic conditions (Sesé et al., 2017). ITGB3 is also associated with acute coronary syndromes (Damar & Eroz, 2020). HIF1α activates the TGFβ-SMAD3 pathway, and both play a critical role in the hypoxic response and response to injury (Basu et al., 2011). SMAD3 is also regulated by serotonin (Chen et al., 2014) which has been implicated in SUID (Cummings & Hodges, 2019). Variants in this gene have been described in patients that died of sudden cardiac death, heart failure and aortic aneurysm (Engström et al., 2020; Hanna et al., 2021; Humeres et al., 2022; Loeys & Dietz, 2008; Ou et al., 2020).
4.2 Potential genetic contributors to SUID vulnerability
Our findings provide further evidence that genetic markers are important indicators for SUID and may contribute to an infant's vulnerability as hypothesized in the triple risk hypothesis (Guntheroth & Spiers, 2002). The number of genetic variations associated with cardiac and neurological disturbances was striking and could contribute to an increased vulnerability during the autoresuscitation response, which critically depends on functional cardiorespiratory coupling and the generation of a hypoxic response. An important consideration that emerges from this and prior genetic studies are the implications for later stages of life. Most individuals with these variants will likely survive the critical period that characterizes SUID but may be at risk for sudden death later in life when faced with other stressors or conditions. For example, we found variants in SCN1A and SCN8A, genes which are known to be associated with SUDEP and have been previously described in SUID, though recognizable seizures had not been observed in these individuals (Chahal et al., 2020; Chahal et al., 2021; Halvorsen et al., 2021; Hammer et al., 2016; Kalume, 2013; Keywan et al., 2021; Koh et al., 2022; Rochtus et al., 2020). We also found many variants in genes associated with cardiac disorders linked to sudden death. For example, Long QT syndrome (CACNA1C, KCNH2) which has been previously implicated in SUID, though these disorders had not yet been observed in these infants (Baruteau et al., 2017; Chahal et al., 2020; Farrugia et al., 2021; Halvorsen et al., 2021; Keywan et al., 2021; Koh et al., 2022; Moore et al., 2020; Opdal & Rognum, 2011; Sutphin et al., 2016; Tester, Wong, Chanana, Gray, et al., 2018; Tester, Wong, Chanana, Jaye, et al., 2018). In addition, we identified affected infants with variants previously reported in ClinVar to be LP/P for conditions such as blood disorders (ALB, HBA2) that have been associated with fatal outcomes, but which were not diagnosed in the infants in which these variants were found (Russo et al., 2019; Toye et al., 2012). These blood disorder variants are especially notable because transition from fetal hemoglobin to that of adults has been implicated in SIDS (Blix, 2020). Adult hemoglobin, which has a lower oxygen affinity, replaces fetal hemoglobin postnatally over a period of six months (Oski, 1979). Variants associated with deafness (GPR98/ADGRV1) are also notable, since inner ear defects have previously been implicated in some infants that succumbed to SUID (Ramirez et al., 2016; Rubens et al., 2008). Several of the variants found in our study were previously reported to be pathogenic for disorders of the muscles including spasticity or parapalegia, (C12orf65/MTRFR, DYNC1H1, PIEZO2, SPG7). Gene enrichment studies aligned with these findings in that notable enrichments in affected infants included motor activity (GO:0003774). Some of these disorders have been associated with respiration dysfunction which could contribute to an individual's vulnerability.
We found many genes associated with the regulation of the hypoxic response and ROS production as mentioned above. HIF-1α and ROS are produced under conditions of chronic intermittent hypoxia (Garcia 3rd et al., 2016), conditions that are seen in apnea of prematurity (Gauda & Master, 2018) and in recurrent apneas thought to precede SUID in some infants. ROS production is a major driver of dysautonomia and could contribute to an abnormal hypoxic response and failed arousal. Disturbances in the regulation of HIF1α and ROS production could exaggerate the accumulation of ROS in response to conditions like intermittent hypoxia thereby increasing the vulnerability of the child to succumb to an exogenous stressor experienced, for example, under prone conditions. This is consistent with pathological discoveries made in children that succumbed to SIDS (Poulsen et al., 1993; Rognum & Saugstad, 1991; Stoltenberg et al., 1993). As previously discussed, the identification of a variant is not sufficient to predict how a variant affects the hypoxic response at the neuronal level, let alone at the level of the child (Opdal & Rognum, 2004). Follow-up studies that generate these human variants in animal models or in IPSCs or human organoids could provide important additional mechanistic clues. Without these functional insights, these discoveries provide novel evidence that aside of the known cardiac, neurological, and metabolic genes, it will be important to consider also genes associated with the regulation of the hypoxic response as potential biomarkers for SUID. However, at this point it is too early to consider certain variants or groups of variants as biomarkers for SUID.
Statistical analysis of genes and pathways provided novel biological insight. For example, the F2R gene contained a splice site variant that was detected in six infants, including both individuals of European and African descent, but only one healthy adult. While primarily a coagulation factor, F2R has also been found to block neuronal apoptosis (Guo et al., 2004), which has been implicated in SUID, particularly in the medulla (Ambrose et al., 2019). Variants in both F2R and HSF1 that were more frequent in affected infants than healthy adults were responsible for the significance of GO:0043280 (positive regulation of cysteine-type endopeptidase activity involved in apoptotic process) in our analysis. Another finding of note was that SUID infants were more likely than the healthy adults to have functional variants in mucin genes. Disruptions to mucin genes could lead to pathogen susceptibility and/or respiratory difficulties (Pinzón Martín et al., 2019). These variants found through statistical analysis, however, were not included here because they did not pass our rigorous filtering, which excluded all variants found in healthy adults. The absolute exclusion of these variants might not necessarily make sense when investigating such a complex disorder as SUID. The triple risk hypothesis suggests that there will be many infants with similar genetic vulnerabilities, not exposed to an exogenous stressor, that will survive infancy, so perhaps excluding variants found in healthy adults or that are found more commonly in a population is not the best approach when examining these infants. Two notable examples of this are demonstrated in this cohort among genes involved in the immune and/or inflammatory response and in serotonin binding and reception. Previous studies have implicated genes involved in these functions in SIDS/SUID (Haynes et al., 2023; Opdal & Rognum, 2011; Opdal, 2018). In our cohort, 72 infants (50.0%) had at least one variant in 31 genes related to immune function or the inflammatory response that have been previously investigated in SIDS (Opdal & Rognum, 2011; Opdal, 2018). Twelve infants had multiple variants in these genes. In all, 76 variants were found in these 31 genes. Nine variants were found in multiple infants, one was found in four infants. None of these variants were found in controls. Seventeen were associated with conditions either inherited in an AD mode or the infants were homozygous for the variant. All were reported in gnomAD at an AF of <0.05% in the total population. However, only two of these variants met our criteria for the predictive computational models (>50% deleterious) and were retained (Table 3). Additionally, 36 children (25.0%) were found with variants in genes involved in serotonin binding and reception which have been implicated previously in SUID (Haynes et al., 2023; Opdal & Rognum, 2011). However, none of these variants made it past our filtering mostly because they were found more frequently in the overall population and thus excluded. It is likely that some of these excluded variants contributed to the overall vulnerability of the infant. In addition, these findings lend support to the idea that genetic vulnerability to SIDS/SUID occurs in a polygenic mode of inheritance for at least a proportion of infants (Opdal & Rognum, 2011). This filtering approach is a possible limitation of this study. New methods of analysis are needed to overcome these deficiencies. Machine Learning and Artificial Intelligence strategies that will accommodate the unbiased analysis of genetic information offer the most promise. Through these approaches, important polygenic interactions and patterns in the data might be uncovered that are missed by current filtering practices.
4.3 Study limitations
We were able to establish demographic aspects such as race/ethnicity, biological sex, and age at death for all but a small proportion of the cohort. For the majority of infants, 87.5%, there were at least partial ME notes describing details surrounding the death scene and/or autopsy results. These details were used to determine the extrinsic and vulnerability risk factors for each infant. However, we were not able to completely characterize the cohort regarding attributes such as the presence of recent illness, medical history, environmental exposures, or even the sleeping circumstances at the time of death due to the partial nature of the notes. The autopsies for 102 of the infants (70.8% of the cohort) received from the NIH NBB were performed by the same medical examiner. For 18 of these, there were no notes beyond race, sex, age at death, and cause of death. This information helps to identify important phenotypic characteristics of an individual and gives investigators insight into their genetic profile. Detailed phenotypic characteristics are highly valuable for establishing a potential causation regarding the genotype of an individual. In addition, parental data was not available for this analysis, and variants were assumed to be de novo. Analysis of trios where this could be confirmed would further strengthen the study.
Mismatched racial proportions between the healthy adults and affected infants are another possible limitation. For the gene level analysis, we demonstrated that significant genes were not confounded by racial differences in the number of variants present (Figure 5). At the variant level, this discrepancy was addressed by matching the appropriate allele frequency in gnomAD specific to the race/ethnicity of the infant in which candidate alleles were identified.
Another possible limitation of this study is the control group comprised of healthy adults enriched in centenarians. It was difficult to find an optimal control group for this study given that every control group comes with certain caveats. A very good control group would have been infants that survived their first year of life. However, the authors are not aware of whether this dataset exists. Moreover, some of these infants could carry the same genes as those that died of SUID, but survived because they did not face other risk factors, such as prone sleeping or smoking. Thus, important genes that increase the vulnerability in combination with other risk factors would be lost. Another control group consisting of infants that died of known causes such as complications of preterm birth or syndromic conditions, would not be ideal as there is likely to be genetic overlap in these conditions with SUID. This would also make it difficult to detect differences and increase the probability that important variants might be missed. It is possible that an infant with the same genetic variant associated with preterm birth or SUID will survive the first year of life due to the absence of an exogenous stressor such as prone sleeping and therefore the death would not be ruled SUID. Including such a child would impact the results of a genetic study, and potentially cause an important genetic variant to be overlooked. This is minimized by making the comparison with healthy adults, though the authors acknowledge that a bias may be introduced. The healthy adults are less likely to have pathogenic variants than the rest of the population, so these variants are more likely to be identified in our cohort due to the filtering process which emphasized variants absent in controls. This bias however, should not present a problem for the candidate gene study that was performed here, but enhanced the identification of these pathogenic variants.
4.4 Conclusion
The present study is one of the first whole genome sequencing studies of infants that died suddenly and unexpectedly. Consistent with the triple risk hypothesis, we find numerous gene variants that could contribute to an increased vulnerability to sudden death. Interestingly, many of those genes have previously been implicated in other causes of sudden death including cardiac death or sudden death in epilepsy. Thus, we hypothesize that many children will survive the critical time period associated with SUID. They will carry this vulnerability into adult life, when they might face other stressors, for example, those known to be associated with sudden cardiac death. This highlights the importance of recognizing vulnerability early in life, which may not only help to prevent SUID, but sudden death in general.
AUTHOR CONTRIBUTIONS
Angela M. Bard: Conceptualization; data curation; formal analysis; investigation; methodology; project administration; supervision; validation; visualization; writing-original draft; writing—review and editing. Lindsay V. Clark: Conceptualization; data curation; formal analysis; investigation; methodology; software; supervision; validation; visualization; writing-original draft; writing—review and editing. Erdal Cosgun: Conceptualization; data curation; formal analysis; investigation; methodology; software; supervision; validation; writing—review and editing. Kimberly A. Aldinger: Conceptualization; formal analysis; investigation; methodology; supervision; validation; writing—review and editing. Andrew Timms: data curation; formal analysis; investigation; software; supervision; validation; writing—review and editing. Lely A. Quina: investigation; methodology; validation. Juan M. Lavista Ferres: investigation; resources; software; validation. David Jardine: investigation; methodology; validation. Elisabeth A. Haas: investigation; methodology; validation. Tatiana M. Becker: project administration; validation. Chelsea M. Pagan: project administration; validation. Avni Santani: data curation; validation; writing—review and editing. Soumitra Barua: data curation; validation. Diego Martinez: data curation; validation. Zakkary McNutt: data curation; validation. Addie Nesbitt: data curation; validation. Edwin A. Mitchell: validation; writing—review and editing. Jan-Marino Ramirez: Conceptualization; formal analysis; funding acquisition; investigation; resources; supervision; validation; visualization; writing-original draft; writing—review and editing.
ACKNOWLEDGMENTS
We would like to thank John and Heather Kahan for their great efforts, financial support, and inspiration to conduct this study.
FUNDING INFORMATION
This study was funded by the Aaron Matthew SIDS Research Guild, Seattle Children's Research Institute, and National Institute of Health (NIH) Grants R01 HL126523 (awarded to Jan-Marino Ramirez), P01 HL090554 (awarded to Jan-Marino Ramirez).
CONFLICT OF INTEREST
The authors declare that they have no interests, financial or otherwise, related to this study that would influence their objectivity or the content of this manuscript.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.




