De novo missense variants in ZBTB47 are associated with developmental delays, hypotonia, seizures, gait abnormalities, and variable movement abnormalities
[Correction added after first online publication on 27 January 2024. The affiliation 8 has been removed and the affiliations 9–13 have been renumbered to 8–12.]
Abstract
The collection of known genetic etiologies of neurodevelopmental disorders continues to increase, including several syndromes associated with defects in zinc finger protein transcription factors (ZNFs) that vary in clinical severity from mild learning disabilities and developmental delay to refractory seizures and severe autism spectrum disorder. Here we describe a new neurodevelopmental disorder associated with variants in ZBTB47 (also known as ZNF651), which encodes zinc finger and BTB domain-containing protein 47. Exome sequencing (ES) was performed for five unrelated patients with neurodevelopmental disorders. All five patients are heterozygous for a de novo missense variant in ZBTB47, with p.(Glu680Gly) (c.2039A>G) detected in one patient and p.(Glu477Lys) (c.1429G>A) identified in the other four patients. Both variants impact conserved amino acid residues. Bioinformatic analysis of each variant is consistent with pathogenicity. We present five unrelated patients with de novo missense variants in ZBTB47 and a phenotype characterized by developmental delay with intellectual disability, seizures, hypotonia, gait abnormalities, and variable movement abnormalities. We propose that these variants in ZBTB47 are the basis of a new neurodevelopmental disorder.
1 INTRODUCTION
Neurodevelopmental disorders are characterized by childhood onset and impairment in personal, social, academic, and occupational functioning. Their presentation includes a variable phenotype of intellectual disability, developmental delay, language and speech disorder, autism spectrum disorder, attention-deficit/hyperactivity disorder, and motor and movement disorder (Neurodevelopmental Disorders, 2013). The identification and understanding of the genetic basis of neurodevelopmental disorders and neurologic diseases are rapidly expanding. The broadening accessibility and growing clinical use of next-generation sequencing (NGS) have resulted in the identification of numerous new disease genes associated with neurodevelopmental phenotypes, including genes encoding transcription factors. There is mounting evidence of the critical role played by transcription factors in the growth and development of the brain, particularly in the differentiation of neural stem cells and the downstream delineation of central nervous system structures (Al-Naama et al., 2020; Lein et al., 2017; Santiago & Bashaw, 2014; Silbereis et al., 2016).
Zinc finger protein transcription factors (ZNFs) make up the largest family of transcription factors in eukaryotes (Al-Naama et al., 2020; Fedotova et al., 2017), including humans (Ladomery & Dellaire, 2002), and serve to regulate cell development and differentiation. They are characterized structurally by finger-like protrusions that bind directly to their related DNA (or other macromolecular) sequence with a zinc ion that creates structural stability through an ionic bond with cysteine or histidine residues of the finger (Al-Naama et al., 2020; Cassandri et al., 2017). Cys2His2 (C2H2) is the most prevalent and widely characterized zinc finger domain. On binding to the targeted DNA sequence, ZNFs containing a C2H2 domain (C2H2-ZNFs) recruit cofactors and other transcription factors to regulate downstream transcription of the targeted gene (Al-Naama et al., 2020). There is evidence that ZNFs are involved in multiple categories of human disease, including immune regulation (Fu & Blackshear, 2017; Maeda & Akira, 2017; Scott & Omilusik, 2019), cancer (Cassandri et al., 2017; Jen & Wang, 2016), skeletal development (Funato et al., 2020), and cardiac disease (Cassandri et al., 2017; Reamon-Buettner & Borlak, 2005). There is also increasing evidence that defects in ZNFs may be associated with neurodevelopmental disorders, neuropsychiatric disorders, and neurological diseases. At least 68 ZNF genes have been reported in association with brain development, neurodevelopmental disabilities, and/or other neuropsychiatric disorders (Al-Naama et al., 2020).
Here we present five unrelated patients with a neurodevelopmental phenotype that includes global developmental delay, intellectual disability, hypotonia, seizures, gait abnormalities, and, in some, abnormal movements unrelated to seizure. Exome sequencing (ES) in all five patients revealed a heterozygous de novo missense variant in the gene ZBTB47 (also known as ZNF651), which encodes a C2H2-ZNF in which variants have not yet been associated with a human phenotype.
2 MATERIALS AND METHODS
2.1 Editorial policies and ethical considerations
This study was approved by ethics committees at the respective institutions involved. All patients or their parents provided written signed consent under a research protocol that was approved by an Institutional Review Board (IRB) at the National Institutes of Health for Patient 1 (P1), Baylor College of Medicine (BCM, P2, P3), UT Health San Antonio (P5), or the Department of Clinical Genetics at Erasmus MC in Rotterdam (in agreement with Dutch research legislation, P4).
2.1.1 Ascertainment
Following the identification of the de novo ZBTB47 candidate variant in P1, additional patients with ZBTB47 variants were identified through case-matching efforts, including responses to P1's Participant Page on the Undiagnosed Diseases Network (UDN) website (http://undiagnosed.hms.harvard.edu/), connection through GeneMatcher (Sobreira et al., 2015), and collaboration with clinical genetic testing laboratories (GeneDx).
2.2 Exome analysis methods
2.2.1 P1
Trio ES was performed on a clinical basis by a commercial clinical genetics laboratory using previously published methods (Yang et al., 2014). The clinical exome data were transferred to the Undiagnosed Diseases Network (BCM site) for research reanalysis, which was performed using Codified Genomics variant review software. De novo and biallelic variants with frequency <1% in gnomAD (Karczewski et al., 2020) and in BCM's local exome database were prioritized for review. Copy number analysis was performed using single nucleotide polymorphism (SNP)-based chromosomal microarray (CMA) at a commercial clinical genetics laboratory.
2.2.2 P2, P3, P5
ES (trio in P2, P5, and proband in P3) was performed on a clinical basis by a commercial clinical laboratory with protocols previously described (Sacoto et al., 2020), and variants reported were confirmed by an orthogonal method, as appropriate. No further exome data analysis beyond the clinical report was performed. Copy number variant analysis via CMA was performed at a commercial clinical genetics laboratory.
2.2.3 P4
Trio ES was performed on a clinical basis by Erasmus MC University Medical Center Department of Clinical Genetics. DNA was enriched using Agilent SureSelect DNA + SureSelect OneSeq 300 kb CNV backbone + Human All Exon V7 capture and paired-end sequenced on the Illumina platform. The data were demultiplexed with bcl2fastq Conversion Software from Illumina. Reads were mapped to the genome using the Burrows-Wheeler Alignment Tool (BWA-MEM algorithm). Sequence variant detection was performed by the Genome Analysis Toolkit HaplotypeCaller. The detected sequence variants were filtered and annotated with Alissa Interpret software and classified with Alamut Visual. Copy number variant analysis via CMA (IL-C12_NEXUS) was performed by Erasmus MC University Medical Center Department of Clinical Genetics.
3 RESULTS
3.1 Case presentations
3.1.1 P1
P1 was a 2-year, 5-month-old female at the time of evaluation by the UDN and 5 years old at her most recent clinical evaluation. Pregnancy was complicated by possible placenta previa per maternal report but without known infection or teratogen exposure. Delivery was via Cesarean section at full term and uncomplicated, and the patient was discharged home in the first few days of life. Family history is unremarkable. Developmental delay was first noticed at 6 months of age, when the patient was unable to sit unsupported and did not respond to sound, reach for objects, or make eye contact. At 8 months she began to demonstrate myoclonic spasms, characterized by flexion of the trunk and upper extremities with head falling forward. Shortly afterward, she began to demonstrate constant movement of hands and feet. At the most recent evaluation, she was unable to sit unassisted or bear weight, but she was able to roll over. She vocalized and babbled but did not have any words. She demonstrated some eye tracking but did not fix and follow, make eye contact, or respond to sounds or commands. She ate by mouth, but gastric tube placement was being considered. At 5 years of age, no significant dysmorphic features were present on physical examination. Musculoskeletal examination and imaging were consistent with mild neuromuscular scoliosis and neuromuscular hip dysplasia. Growth parameters remained in the normal range (between 20–50th percentile for weight and between 30–80th percentile for height) until around 4 years of age, at which time she began to demonstrate weight loss and poor growth. Most recent growth parameters were as follows: less than the 0.01st percentile for weight (Z-score: -4.09), 0.58th percentile for height (Z-score: -2.53), and 11th percentile for occipital frontal circumference (OFC, Z-score: -1.23). She was evaluated by gastroenterology (GI), diagnosed with severe malnutrition, and started on caloric supplementation. She was referred to neurology due to concern for significant developmental delay, abnormal movements, and seizure activity. On neurological examination, she demonstrated significant generalized hypotonia and near continuous choreiform movements of hands, feet, trunk, and face. She was started on anti-epileptic drugs (AEDs) with poor control of her seizures and continued to have approximately 45 seizures per day. Most recent epilepsy management included clobazam and cannabidiol, with consideration of starting a modified Atkins diet.
Electroencephalogram (EEG) showed multifocal spikes and generalized tonic seizures. Multiple magnetic resonance imaging (MRI) studies of the brain were normal, as was MR spectroscopy. Audiology examinations via otoacoustic emissions and auditory brainstem response were normal. Cerebrospinal fluid (CSF) neurotransmitters, CSF lactate, serum lactate, ammonia, and pyruvic acid were all normal. Metabolic studies were within normal limits, including plasma amino acids, acylcarnitine analysis, and urine organic acids. NMDA, thyroglobulin, and GAD antibodies were negative. Metabolomic studies of the CSF and plasma were performed and were remarkable for elevated plasma pipecolic acid, but other metabolites associated with peroxisomal disorders were unremarkable. CMA was normal. Angelman syndrome methylation analysis and myotonic dystrophy type 1 repeat expansion analysis were normal/negative. Sterol studies for investigation of cholesterol synthesis disorders were normal. Clinical trio exome sequencing (ES) was performed and identified a heterozygous variant of uncertain significance (VUS) in CHRNB2 inherited from the unaffected mother. Additional details on the CHRNB2 variant as well as other variants reported are provided in Table S1. CHRNB2 is associated with nocturnal frontal lobe epilepsy 3. ES was otherwise considered non-diagnostic clinically with no pathogenic, likely pathogenic variants, or de novo VUS in known disease genes. The patient was referred to the UDN for further evaluation, and research reanalysis of the ES data highlighted a heterozygous de novo variant in ZBTB47 [NM_145166.3:c.2039A>G, p.(Glu680Gly)] as a candidate of interest.
3.1.2 P2
P2 was an 11-month-old female at initial presentation and 6 years old at the most recent clinical evaluation. Pregnancy history was complicated by preeclampsia. The patient was born at 37 weeks gestational age via vaginal delivery and was transferred to the neonatal intensive care unit (NICU) briefly due to respiratory difficulties. She was discharged at 2 days of life. Breathing and feeding difficulties were present since birth. The patient was diagnosed with laryngomalacia, laryngeal cleft, reflux, and dysphagia. She underwent supraglottoplasty with improvement in respiratory disease. The patient also underwent G-tube placement due to dysphagia and growth concerns. The patient's history was remarkable for hypotonia and global developmental delays. She sat at 13 months and walked at 18 months. Her first words were reported at 2.5 years of age, and two-word phrases were noted at 4 years. The patient also demonstrated fine motor delay. At the most recent evaluation, she continued to undergo speech, occupational, and physical therapy twice per week. Physical examination demonstrated dysmorphic features including tented mouth, deep-set eyes, high-arched palate, small nose, relative microcephaly, wrinkly skin, and sagging cheeks. Most recent growth parameters were as follows: 69th percentile for height (Z-score: +0.50), 41st percentile for weight (Z-score: -0.23), and 4th percentile (Z-score: -1.75) for head circumference. The patient was diagnosed with generalized seizures at 2 years of age. Seizures presented during sleep as startling/shuddering attacks. The patient was most recently on valproic acid but continued to have 2–4 seizures per month. She also continued to startle easily. The neurological examination was remarkable for Gowers sign, ataxic gait, diffuse weakness, hypotonia, myoclonus, and hand tremor. Due to developmental and behavioral concerns (tantrums and poor sociability), she received neuropsychological evaluation and was found to have moderate intellectual disability and autism spectrum disorder. Other health concerns of note included muscular ventricular septal defect (VSD), frequent infections, sleep apnea, gastroesophageal reflux disease (GERD), constipation, pressure equalization (PE) tubes, and the requirement of glasses.
Brain MRI was normal. EEG showed primary generalized epilepsy with occasional runs of bifrontal predominant generalized spike and wave discharges with shifting bilateral predominance, occipital intermittent rhythmic delta activity (OIRDA), and occasional irregular and rhythmic delta activity in left greater than right hemispheres. Genetic evaluation included SNP-CMA, which revealed small regions of homozygosity (<10 Mb) but was otherwise unremarkable, Prader-Willi methylation studies that were normal, and DMPK sequencing that was negative. Trio ES with mitochondrial sequencing identified a heterozygous VUS in SLC12A5 [NM_020708.4:c.1955C>T, p.(Ala652Val)] inherited from the unaffected mother. Additional details on this variant may be found in Table S2. This gene is associated with autosomal dominant susceptibility to idiopathic generalized epilepsy and autosomal recessive developmental and epileptic encephalopathy. No other variants were reported. ES reanalysis 2 years later identified a heterozygous de novo variant in ZBTB47 [NM_145166.3:c.1429G>A, p.(Glu477Lys)].
3.1.3 P3
P3 was a 5-year-old male on initial evaluation and 8 years old at the most recent clinical evaluation. Pregnancy was complicated by maternal hypothyroidism treated with levothyroxine. There was no known infection or teratogen exposure during pregnancy. Delivery was vaginal, full-term, and uncomplicated. Family history was unremarkable. Weight at birth was 3.572 kg (51st percentile for age, Z-score: +0.03); length at birth was 48.3 cm (24th percentile for age, Z-score: -0.71). The neonatal period was uncomplicated. The patient walked at 18 months. His first words occurred at 2 years after receiving speech therapy, and he started using two-word phrases at 5 years. Early language was dominated by echolalia. At the most recent evaluation he remained unable to read or write and did not know letters or numbers. His behavior was remarkable for frequent tantrums, screaming with frustration, poor eye contact, and poor sociability. His neuropsychological evaluation was notable for behavioral dysregulation, language and cognitive impairment, fine motor difficulties, and moderate intellectual disability. He was diagnosed with autism spectrum disorder. At the most recent evaluation he was feeding by mouth and demonstrated poor growth with the following parameters: weight at the 1st percentile for age (Z-score: -2.47), height at the 5th percentile for age (Z-score: -1.66), and OFC at the 74th percentile for age (Z-score: +0.64). Physical examination demonstrated dysmorphisms, including prominent forehead, deep orbits, borderline hypertelorism, and anteverted nose. Neurological examination was remarkable for hand tremor, wide-based gait, and poor coordination that worsened with time. He was also affected by partial seizures with onset at 17 months old that remained refractory to AEDs (2–4 seizures per month), and he continued off AEDs. Of note, the patient's history was also remarkable for frequent infections, including recurrent otitis media, Salmonella gastroenteritis, and sepsis that required hospitalization. Other medical problems included mild hearing loss, sleep apnea, constipation, intermittent tachycardia, joint laxity, and aortic valve abnormality. Surgical history included tonsillectomy, adenoidectomy, and bilateral PE tube placement.
EEG was remarkable for independent left and right parietal epileptiform discharges. MRI of the brain demonstrated cortical dysplasia of the left temporal lobe and Chiari I malformation, as well as haziness of the gray-white matter interface at the temporal poles. The positron emission tomography (PET) of the brain was consistent with decreased glucose metabolism in the temporal lobes and left frontal lobe. Serum lactate was below normal, and 2-hydroxyglutaric acid and acetoacetic acid were elevated, but these findings were considered nonspecific and non-diagnostic. CMA was normal. Proband ES identified a heterozygous paternally inherited VUS in PPOX (NM_003009.5:c.338+2dupT). Additional details on this variant may be found in Table S3. Heterozygous pathogenic variants in PPOX are associated with porphyria variegata. The variant observed in this patient has been previously reported as likely pathogenic in association with this disorder (Rossetti et al., 2008). Pathogenicity is supported by the variant's location at a consensus splice site. However, this variant is paternally inherited from a currently unaffected father suggesting the possibility of incomplete penetrance or benign status. In addition, porphyria variegata was determined to be an incomplete phenotype match for the patient. A heterozygous variant in ZBTB47 was also detected [NM_145166.3:c.1429G>A, p.(Glu477Lys)] and was determined to be de novo after parental testing. No other variants were reported.
3.1.4 P4
P4 was a 3-year-old female at first evaluation and 13 years old at the most recent clinical evaluation. Pregnancy was complicated by gestational diabetes and prolonged rupture of membranes. The patient was born at 38 weeks gestation. Weight at birth was 2.690 kg (8th percentile for age; Z-score: -1.4); length at birth was 50 cm (58th percentile for age; Z-score: +0.20); OFC at 4 weeks of age was 35.5 cm (16th percentile for age; Z-score: -0.99). The neonatal period was uncomplicated. Family history was remarkable for a paternal half-aunt with epilepsy that presented after a case of meningitis. The patient's development was relatively unremarkable early in life with some indication of gross motor delay. She sat unassisted at 5 months, crawled at 10 months, and walked at 16 months. She spoke her first word at 12 months and was toilet trained by 18 months. Her developmental delay became more apparent with age. By the age of 2 years, she was noted to have some difficulty learning. At 3 years old she was noted to have stiff movements with poor gross motor skills and was unable to speak in three-word phrases. She was diagnosed with moderate intellectual disability at 7 years of age, and she received a diagnosis of autism spectrum disorder, characterized by poor eye contact, echolalia, difficulty with social interaction, difficulty accommodating change, anger outbursts, and anxious behavior. The patient was also diagnosed with epilepsy. She was first noted to have staring episodes around 2–3 months of age, followed by febrile seizures at 13 months and 3 years and recurrent tonic–clonic seizures with multiple episodes of status epilepticus starting at age 5 years. She continued to have severe and refractory multifocal epilepsy at the most recent evaluation with frequency of 12 seizures per month. Her seizures were managed with clobazam, lamotrigine, midazolam, and sulthiame with improvement. Her most recent growth parameters were as follows: weight at the 58th percentile (Z-score: +0.21), height at the 87th percentile (Z-score: +1.12), and OFC at the 32nd percentile (Z-score: -0.47). Physical examination was notable for dysmorphic features including deep orbits, full cheeks, prominent forehead, small bitemporal diameter, and full upper eyelids. Neurological examination was also remarkable for hypotonia, generalized hypermobility, unsteady gait, poor coordination, and hand-flapping with excitement. The patient's medical history was otherwise notable for chronic constipation.
EEG demonstrated multifocal epileptic pattern with consistent focus in the left centroparietal region. Brain MRI was notable only for slightly wide occipital horns. Non-diagnostic genetic testing included epilepsy gene panel, intellectual disability gene panel, SCN1A sequencing, PCDH19 sequencing, CDKL5 sequencing, mitochondrial DNA sequencing, and CMA. Trio ES identified a heterozygous de novo variant in ZBTB47 [NM_145166.4:c.1429G>A, p.(Glu477Lys)]. Additional variants identified may be found in Table S4.
3.1.5 P5
P5 was a 6-year-old male at evaluation. Pregnancy was uncomplicated, and he was born at full term via vaginal delivery. Weight at birth was 3.529 kg (64th percentile; Z-score: +0.36); length at birth was 53 cm (89th percentile; Z-score: +1.20); OFC at birth was 34.5 cm (50th percentile; Z-score: 0.0). His neonatal period was complicated by a brief NICU hospitalization for transient tachypnea of the newborn. Family history was remarkable for febrile seizures in the patient's father and autism spectrum disorder in a paternal first cousin. The patient's infancy was complicated by global developmental delay. He sat unassisted at 7 months of age and walked at 15 months of age. He continued to walk with an unsteady gait at the most recent evaluation. His first words occurred at 2 years old, and at the most recent evaluation he had only 30–40 words with noted echolalia. His receptive language was better than his expressive language, and he was diagnosed with autism spectrum disorder. He began having seizures at 9 months of age and was diagnosed with generalized tonic–clonic epilepsy with focal motor seizures. At the most recent evaluation his seizures were controlled on lacosamide and valproic acid with breakthrough seizures noted every 2–3 months. He was also noted to have a tic disorder and hyperactivity. His most recent growth parameters were as follows: weight at the 83rd percentile (Z-score: +0.95), height at the 98th percentile (Z-score: +2.22), OFC at the 14th percentile (Z-score: -1.08). Physical examination was notable for dysmorphic features including cupped ears, short philtrum with thick upper lip, and small café au lait macule on the left thigh. The patient's neurological examination was also remarkable for hypotonia. Other medical problems included dysphagia and chronic rhinitis.
EEG was consistent with the patient's clinical epileptic activity. MRI of the brain was normal. Metabolic workup, including plasma amino acids, urine organic acids, acylcarnitine profile, lactic acid, and pyruvate, was normal. Further genetic workup was also non-diagnostic, including CMA, fragile X testing, an autism/intellectual disability gene panel, and an epilepsy gene panel. Trio ES performed during a genetics reevaluation identified a heterozygous de novo variant in ZBTB47 [NM_145166.3:c.1429G>A, p.(Glu477Lys)]. No other variants were reported.
3.2 Variant interpretation
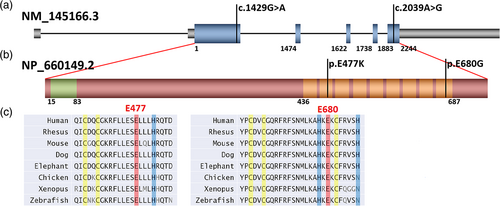
P1 is heterozygous for a de novo p.(Glu680Gly) (c.2039A>G) missense variant in ZBTB47. This variant localizes to the last exon (exon 6) of ZBTB47 and the final zinc finger domain of the ZBTB47 protein (Kopanos et al., 2019; The UniProt Consortium, 2021) (Figure 1). Analysis of the exome data from P2, P3, P4, and P5 revealed the same de novo heterozygous p.(Glu477Lys) (c.1429G>A) missense variant in ZBTB47. This variant localizes to exon 2 of ZBTB47 and the second zinc finger domain, which is noted to be degenerate (Kopanos et al., 2019; The UniProt Consortium, 2021) (Figure 1). Of note, the UniProt database annotates nine total zinc finger domains in ZBTB47 (The UniProt Consortium, 2021) (Figure 1), while the original paper characterizing ZBTB47 only noted seven zinc finger domains (Kumar et al., 2010), excluding the two where the patients' variants reside, suggesting that these regions less stringently correlate with a canonical C2H2 zinc finger sequence. Despite residing in less typical zinc finger domains, the bioinformatics of both variants support pathogenicity (Table 1). Neither variant was present in the gnomAD (Karczewski et al., 2020) or ClinVar databases. The GERP scores, a measure of conservation at nucleotide position, are 4.5799 and 4.28, respectively (Cooper et al., 2005), which demonstrate a high level of conservation at these positions. Amino acid conservation is also high across species at both loci (The UniProt Consortium, 2021; Figure 1c). The combined annotation-dependent depletion (CADD) scores, a measure of the deleteriousness of variants, are high at 29.9 and 25.3, respectively, and pathogenicity prediction models SIFT and PolyPhen report that both variants are “deleterious” and “probably damaging,” respectively.
| Patient 1 (P1) | Patient 2 (P2) | Patient 3 (P3) | Patient 4 (P4) | Patient 5 (P5) | |
|---|---|---|---|---|---|
| Variant (NM_145166.3) | c.2039A>G, p.(Glu680Gly) | c.1429G>A, p.(Glu477Lys) | |||
| Position (GRCh37/hg19) | chr3:42705885 | chr3:42701276 | |||
| Exon number (total = 6 exons) | 6 | 2 | |||
| Inheritance | De novo | De novo | |||
| Testing | Trio exome sequencing with Sanger confirmation (Baylor Genetics) | Trio exome sequencing (GeneDx) | Exome Sequencing (GeneDx) with subsequent parental testing | Trio exome sequencing (Erasmus MC University Medical Center Department of Clinical Genetics) | Trio exome sequencing (GeneDx) |
| Present in ClinVar | Previously unreported | Previously unreported | |||
| Present in gnomAD | No | No | |||
| GERP | 4.58 | 4.28 | |||
| CADD | 29.9 | 25.3 | |||
| SIFT | Deleterious | Deleterious | |||
| PolyPhenCat | Probably damaging | Probably damaging | |||
| REVEL | Uncertain (0.5) | Uncertain (0.35) | |||
| ACMG criteria | PS2, PM2, PP3 | PS2, PM2, PP3 | |||
4 DISCUSSION
Over 68 ZNF genes have been reported in association with brain development, neurodevelopmental disabilities, and/or other neuropsychiatric disorders (Al-Naama et al., 2020). ZNFs reported in association with developmental delay, autism spectrum disorder, and/or seizures include ZEB2 (Baxter et al., 2017; Buraniqi & Moodley, 2015; Ghoumid et al., 2013; Yuan et al., 2015), BCL11A (Dias et al., 2016; Shimbo et al., 2017; Yoshida et al., 2018), PLZF (Fischer et al., 2008), ZBTB11 (Fattahi et al., 2018), ZNF292 (Mirzaa et al., 2020), ZBTB18 (Depienne et al., 2017; van der Schoot et al., 2018), ZNF462 (Cosemans et al., 2018; Kruszka et al., 2019; Weiss et al., 2017), ZNF778 (Willemsen et al., 2010), ZNF41 (Shoichet et al., 2003), ZNF711 (van der Werf et al., 2017), ZBTB20 (Cordeddu et al., 2014; Jones et al., 2018; Mattioli et al., 2016), ZNF407 (Kambouris et al., 2014), ZNF148 (Stevens et al., 2016), ZNF142 (Khan et al., 2019), TSHZ3 (Caubit et al., 2016), GLI3 (Biesecker, 2006), ZDHHC8 (Yang et al., 2018), ZNF81 (Kleefstra et al., 2004), ZNF804A (Anitha et al., 2014), and ZNF674 (Lugtenberg et al., 2006). Other reports document copy number variants, which encompass multiple ZNFs and a neurodevelopmental phenotype including developmental delay and seizures (Andrieux et al., 2008; Spreiz et al., 2014). To date, ZBTB47 has not been associated with human disease. In this report, we present five patients with one of two de novo heterozygous missense variants in ZBTB47 with bioinformatics supportive of pathogenicity (Table 1) and a phenotype that includes developmental delay, intellectual disability, seizures, hypotonia, and variable abnormal movements (Table 2).
| Patient 1 (P1) | Patient 2 (P2) | Patient 3 (P3) | Patient 4 (P4) | Patient 5 (P5) | |
|---|---|---|---|---|---|
| Age | 5y | 6y | 8y | 13y | 6y |
| Sex | F | F | M | F | M |
| Pregnancy history | Placenta previa; Full term | Preeclampsia; Born at 37 weeks; Pneumothorax requiring no intervention | Maternal hypothyroidism treated with levothyroxine; Full term | Gestational diabetes, prolonged rupture of membranes; Born at 38 weeks | No complications; Full term |
| Birth weight (kg; Z-score) | Unknown | Unknown | 3.572; 0.03 | 2.690; −1.4 | 3.529; 0.36 |
| Birth length (cm; Z-score) | Unknown | Unknown | 48.3; −0.71 | 50; −0.71 | 53; 1.20 |
| Birth OFC (cm; Z-score) | Unknown | Unknown | Unknown | Unknown | 34.5; 0.00 |
| Weight (Z-score) | −4.09 | −0.23 | −2.47 | +0.21 | +0.95 |
| Height (Z-score) | −2.53 | +0.50 | −1.66 | +1.12 | +2.22 |
| OFC (Z-score) | −1.23 | −1.75 | +0.64 | −0.47 | −1.08 |
| Dysmorphic features | Non-dysmorphic | Tented mouth, deep-set eyes, asymmetric facies, sagging cheeks | Deep orbits, prominent forehead, anteverted nose, borderline hypertelorism | Deep orbits, full cheeks, small bitemporal diameter | Short philtrum with thick upper lip, cupped ears |
| Developmental delay | Yes | Yes | Yes | Yes | Yes |
| Age at walking | Non-ambulatory | 18 months | 18 months | 16 months | 15 months |
| Age at talking | Nonverbal | 2.5 years | 2 years | 1 year | 2 years |
| Age at putting words together | Nonverbal | 4 years | 5 years | 4.5 years | Unknown |
| Intellectual disability | Unknown | Moderate | Moderate | Moderate | Moderate |
| Hypotonia | Yes | Yes | Yes | Yes | Yes |
| Coordination | Poor | Poor | Poor | Poor | Poor |
| Abnormal behaviors | Teeth grinding | Easy startle, strong shudder reflex | Aggressive behavior, frequent tantrums, poor eye contact, poor sociability | Aggressive behavior, anxious behavior, echolalia | Hyperactivity |
| Autism spectrum diagnosis | No | Yes | Yes | Yes | Yes |
| Seizures | Yes | Yes | Yes | Yes | Yes |
| Seizure age of onset | 8 months | 2 years | 17 months | 2 months | 9 months |
| Seizure type | Refractory tonic–clonic seizures, generalized slowing | Generalized epilepsy, multiple seizure types | Partial seizures, Multifocal parietal epileptiform discharges | Refractory tonic–clonic seizures, Multifocal epilepsy with left centroparietal region focus | Generalized tonic–clonic epilepsy with focal motor seizures |
| Seizure frequency | 45 per day | 2–4 per month | 2–4 per month | 12 per month | 2–3 per month |
| Anti-epileptic drug therapy | Yes – cannabidiol, clobazam | Yes – valproic acid | No | Yes – clobazam, lamotrigine, midazolam, sulthiame | Yes – lacosamide, valproic acid |
| Abnormal movements | Choreiform movements of hands, feet, trunk, and face | Twitching during sleep, shuddering attacks, easy startle, choreiform movements when walking | Hand tremor | Hand-flapping with excitement | Tic disorder |
| Gait abnormalities | Non-ambulatory | Gowers sign | Wide-based gait | Unsteady | Unsteady |
| Cardiovascular abnormalities | Nonspecific ST- and T-wave abnormality | Muscular ventricular septal defect | Intermittent tachycardia, aortic valve abnormality | None, pending evaluation | None |
| Gastrointestinal problems | Severe malnutrition | Constipation, GERD, dysphagia | Poor weight gain, lactose intolerance, constipation | Constipation | Dysphagia |
| Skeletal abnormalities | Scoliosis, hip dysplasia | None | Joint laxity | Hypermobility | None |
| Other medical problems | Frequent viral infectious | Frequent viral infections, laryngomalacia with laryngeal cleft, sleep apnea | Frequent infections, recurrent otitis media, Salmonella gastroenteritis, stridor, sleep apnea, hearing loss | None | Chronic rhinitis |
| MRI results | Normal | Normal | Cortical dysplasia of left temporal lobe, Chiari I malformation, poor definition of gray-white matter interface at temporal poles | Slightly wide occipital horns | Normal |
| Chromosomal microarray | Normal | Regions of homozygosity (<10 Mb) | Normal | Normal | Normal |
| Other non-diagnostic genetic testing | Myotonic dystrophy panel, PWS/AS methylation | Myotonic dystrophy panel, PWS/AS methylation | None | Epilepsy gene panel, intellectual disability panel, mtDNA sequencing, sequencing of CDKL5, PCDH19, SCN1A | Fragile X, autism/intellectual disability panel, epilepsy panel |
- Abbreviations: F, female; GERD, gastroesophageal reflux disease; M, male; OFC, occipital frontal circumference; PWS/AS, Prader-Willi syndrome/Angelman syndrome.
ZBTB47 (also known as ZNF651) is a ZNF652 paralogue that was first described as a classical C2H2-ZNF located on chromosome 3 (Kumar et al., 2010). The currently recognized canonical transcript (NM_145166.3) encodes a 747 amino acid protein, which includes a longer N-terminal region with a BTB domain not recognized in this previous publication. ZBTB47 and ZNF652 demonstrate high conservation in their zinc finger domains (95%) and short carboxy-terminal proline-rich sequence (85%) (Kumar et al., 2010). ZNF652 has been previously demonstrated to interact at its proline-rich carboxy terminus with the CBFA2T3 protein of the ETO family of transcriptional regulatory proteins to repress transcription (Kumar et al., 2008, 2006). ZBTB47 demonstrates binding of the same DNA sites and analogous interaction with CBFA2T3 at its proline-rich region of the carboxy terminus, suggesting it also represses transcription (Kumar et al., 2010). However, tissue expression of the two paralogues is different, suggesting their developmental function may differ. ZBTB47 is widely expressed, with highest expression noted in cardiovascular tissues, skeletal muscle, and central nervous system tissues per GTEx (The GTEx Consortium, 2020), a tool for comparison of protein expression among different tissue types. The probability of being loss-of-function intolerant (pLI) for ZBTB47 is 1.0, the missense Z-score for the gene is 2.45, and the observed overexpected (o/e) missense score is 0.66 (Karczewski et al., 2020), all of which support pathogenicity of our reported missense variants.
The phenotype of our patients is compatible with the general phenotype of neurodevelopmental disorder and developmental delay described in other reported ZNF syndromes but is distinguished by the presence of variable abnormal movements in all patients. P1's movements are choreiform in nature and are seen in the hands, feet, trunk, and face. P2 also has choreiform movements with ambulation, as well as shuddering attacks, twitching during sleep, and easy startling. As a result, choreiform movements are seen in association with both reported variants. P3 does not demonstrate choreiform movements but does present with a significant hand tremor. P4 demonstrates hand-flapping, and P5 presents with a tic disorder. All five patients have poor coordination and abnormal gait. P1 is not ambulatory, P2 demonstrates a Gowers sign in preparation for ambulation, and P3, P4, and P5 walk with a wide-based or unsteady gait. Abnormal movements have been reported in association with ZNF syndromes at least twice previously (Fattahi et al., 2018; Khan et al., 2019), but this finding remains uncommon and may be distinguishing for patients with pathogenic ZBTB47 variants.
Other less specific neurodevelopmental abnormalities are also present in our cohort of patients, including developmental delay, the primary trigger for evaluation in all five patients. Early motor and speech milestones were mildly delayed in most patients with P1 (who remains non-ambulatory and nonverbal) being an exception. Delays in achieving more advanced skills became more obvious with age. Patients experienced difficulty with motor skills throughout childhood, with poor coordination likely contributing, and their ability to put words together and form sentences were more delayed than age at first words. All continue to demonstrate persistent speech delay. Developmental regression has not been seen in our patients. Moderate intellectual disability has been confirmed in P2, P3, P4, and P5, and intellectual disability is suspected in P1. Four of five patients (P2, P3, P4, and P5), all sharing the p.(Glu680Gly) variant, have been diagnosed with autism spectrum disorder. All reported patients have been diagnosed with seizures and demonstrate hypotonia on examination. The characterization and severity of seizures reported vary (Table 2), but the seizures are refractory in two patients (P1 and P4) and incompletely controlled in two (P2 and P3). The reported seizures do not demonstrate consistent response to specific AEDs. MRI was normal for P1, P2, and P5. P3 was found to have a Chiari I malformation on MRI, as well as cortical dysplasia of the left temporal lobes and poor definition of gray-white matter interface at the temporal lobe. P4 was noted to have slightly wide occipital horns on imaging.
Three of five patients demonstrate growth discrepancy. Both P1 and P3 were appropriate for gestational age at birth but have since developed failure to thrive. Both patients are followed by GI and are being evaluated for failure to thrive and malnutrition. P2 has relative microcephaly with head circumference around the 4th percentile but with normal height and weight. No consistent pattern of dysmorphic features is seen across our patients, and there is no clear dysmorphic feature profile to suggest for this disorder at this time, although 4 of 5 patients are noted to demonstrate variable dysmorphic facial features (P2, P3, P4, P5).
There are two other features that stand out as possible distinguishing characteristics when examining the phenotypes of our reported patients: cardiac disease and immune dysregulation. Three patients demonstrate a cardiac abnormality of some kind, including structural anomalies (P2 and P3), arrhythmia (P3), and nonspecific EKG findings (P1). When considering the cardiac phenotypes of these patients in the context of ZBTB47's high expression in cardiovascular tissues and the understanding that other ZNFs are associated with cardiac disease (Cassandri et al., 2017; Reamon-Buettner & Borlak, 2005), the presence of a cardiac disease component of this syndrome is possible. Four of our patients have a history of recurrent or frequent infections, including frequent viral infections (P1 and P2), chronic rhinitis (P5), and recurrent bacterial infection and sepsis (P3). None of the reported patients had abnormal immunological testing. However, a history of frequent infection is difficult to interpret as viral infections are common in children, the frequency and severity of which vary significantly in relation to environmental exposure, health practices, and comorbidities. Nevertheless, the history of infection seen in our patients, along with the knowledge that some ZNFs play a role in immune regulation (Fu & Blackshear, 2017; Maeda & Akira, 2017; Scott & Omilusik, 2019), raises the question of possible immune dysregulation not detected by standard testing in association with this syndrome.
This study is limited in its focus on only five patients. The small size of our current cohort is likely related to the rarity of pathogenic variants in ZBTB47 and the gene's lack of inclusion in most current sequencing platforms. Further limitations include the general paucity of information on the function of ZBTB47, as well as the lack of functional data and absence of animal models to support pathogenicity of these variants on the biochemical or organismal level. Neurodevelopmental phenotypes are generally nonspecific. As such, it is difficult to completely rule out the influence of other disease-causing variants in our patients, although the presence of the same de novo variant in four unrelated cases [c.1429G>A, p.(Glu477Lys)] is supportive of pathogenicity (and suggests this amino acid plays an important role in protein function). In addition, the shared phenotype among our patients, including developmental delay, intellectual disability, hypotonia, seizures, and possible movement abnormalities, is consistent and distinguished enough to attribute to the reported de novo missense variants in ZBTB47.
The mechanism by which these ZBTB47 variants result in the described neurodevelopmental phenotype is unclear and will require further research. Development of the central nervous system is complex, and evidence describing the role that transcription factors play in this process continues to grow (Al-Naama et al., 2020; Gower-Winter & Levenson, 2012; Lein et al., 2017; Santiago & Bashaw, 2014; Silbereis et al., 2016). Biochemically, more work is needed to fully elucidate the function of ZBTB47 and the pathogenesis of the presented variants at the molecular level to allow for a better understanding of the development of the phenotype, as well as consideration of future management options. The mechanism of pathogenicity for these variants is currently unclear, though the pLI score of ZBTB47 is 1.0, suggesting strong intolerance of loss-of-function variants and lending support to a possible hypomorphic or haploinsufficiency mechanism. However, gain-of-function or dominant negative mechanisms are also possible. Further functional studies are needed to establish the mechanism by which these variants cause the phenotype. Future clinical studies could involve the collection of more patients identified with ZBTB47 variants into a more extensive review of phenotype and natural history. In addition, RNA sequencing could prove useful to characterize the impact that these variants have on the expression of other genes given that ZBTB47 is a transcription factor.
In summary, this is the first report of de novo heterozygous variants in ZBTB47 in association with a human phenotype. The heterozygous missense variants detected in five unrelated patients via ES are associated with a variable autosomal dominant neurodevelopmental phenotype including developmental delay and intellectual disability, hypotonia, seizures, gait abnormalities, and variable abnormal movements.
AUTHOR CONTRIBUTIONS
Scott K. Ward involved in conceptualization, methodology, investigation, resources, data curation, writing the original draft, review, editing, and visualization. Alexandrea Wadley and Chun-hui (Anne) Tsai involved in conceptualization, resources, investigation, review, and editing. Paul J. Benke involved in conceptualization, resources, investigation, review, editing, and supervision. Lisa Emrick, Kristen Fisher, William Craigen, and Kimberly Houck involved in resources, investigation, review, and editing. Hongzheng Dai involved in resources, investigation, and data curation. Undiagnosed Diseases Network: Conceptualization, Investigation, Resources. Maria J. Guillen Sacoto involved in resources, review, and editing. Kimberly Glaser, Luis Rohena, Karin E.M. Diderich, and Hennie T. Bruggenwirth involved in conceptualization, resources, investigation, review, and editing. David R. Murdock involved in resources and data curation. Brendan Lee and Carlos Bacino involved in resources, investigation, review, and editing. Lindsay C. Burrage involved in conceptualization, methodology, investigation, resources, data curation, supervision, project administration, review, and editing. Jill A. Rosenfeld involved in conceptualization, methodology, investigation, resources, data curation, visualization, supervision, project administration, review, and editing.
ACKNOWLEDGMENTS
We would like to thank the patients and families for their participation. Research reported in this manuscript was supported by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director under Award Number U01HG007709. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. L.C.B. holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund. This project was supported in part by the Clinical Translational Core (CTC) of the Baylor College of Medicine (BCM) Intellectual and Developmental Disabilities Research Center (IDDRC). The BCM IDDRC is supported by P50 HD103555 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD). The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from: the GTEx Portal on 04/16/21 and/or dbGaP accession number phs000424.vN.pN on 04/16/21.
CONFLICT OF INTEREST STATEMENT
The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing completed at Baylor Genetics Laboratory. MJGS is an employee of GeneDx, LLC.
EDITORIAL POLICIES AND ETHICAL CONSIDERATIONS
This study was approved by ethics committees at the respective institutions involved. All patients or their parents provided written signed consent under a research protocol that was approved by an Institutional Review Board (IRB) at the National Institutes of Health for Patient 1 (P1), Baylor College of Medicine (BCM, P2, P3), UT Health San Antonio (P5), or the Department of Clinical Genetics at Erasmus MC in Rotterdam (in agreement with Dutch research legislation, P4).
APPENDIX A: Members of the Undiagnosed Diseases Network
Maria T. Acosta, Margaret Adam, David R. Adams, Raquel L. Alvarez, Justin Alvey, Laura Amendola, Ashley Andrews, Euan A. Ashley, Carlos A. Bacino, Guney Bademci, Ashok Balasubramanyam, Dustin Baldridge, Jim Bale, Michael Bamshad, Deborah Barbouth, Pinar Bayrak-Toydemir, Anita Beck, Alan H. Beggs, Edward Behrens, Gill Bejerano, Hugo J. Bellen, Jimmy Bennett, Beverly Berg-Rood, Jonathan A. Bernstein, Gerard T. Berry, Anna Bican, Stephanie Bivona, Elizabeth Blue, John Bohnsack, Devon Bonner, Lorenzo Botto, Brenna Boyd, Lauren C. Briere, Gabrielle Brown, Elizabeth A. Burke, Lindsay C. Burrage, Manish J. Butte, Peter Byers, William E. Byrd, John Carey, Olveen Carrasquillo, Thomas Cassini, Ta Chen Peter Chang, Sirisak Chanprasert, Hsiao-Tuan Chao, Ivan Chinn, Gary D. Clark, Terra R. Coakley, Laurel A. Cobban, Joy D. Cogan, Matthew Coggins, F. Sessions Cole, Heather A. Colley, Heidi Cope, Rosario Corona, William J. Craigen, Andrew B. Crouse, Michael Cunningham, Precilla D'Souza, Hongzheng Dai, Surendra Dasari, Joie Davis, Jyoti G. Dayal, Esteban C. Dell'Angelica, Patricia Dickson, Katrina Dipple, Daniel Doherty, Naghmeh Dorrani, Argenia L. Doss, Emilie D. Douine, Dawn Earl, David J. Eckstein, Lisa T. Emrick, Christine M. Eng, Marni Falk, Elizabeth L. Fieg, Paul G. Fisher, Brent L. Fogel, Irman Forghani, William A. Gahl, Ian Glass, Bernadette Gochuico, Page C. Goddard, Rena A. Godfrey, Katie Golden-Grant, Alana Grajewski, Don Hadley, Sihoun Hahn, Meghan C. Halley, Rizwan Hamid, Kelly Hassey, Nichole Hayes, Frances High, Anne Hing, Fuki M. Hisama, Ingrid A. Holm, Jason Hom, Martha Horike-Pyne, Alden Huang, Sarah Hutchison, Wendy Introne, Rosario Isasi, Kosuke Izumi, Fariha Jamal, Gail P. Jarvik, Jeffrey Jarvik, Suman Jayadev, Orpa Jean-Marie, Vaidehi Jobanputra, Lefkothea Karaviti, Shamika Ketkar, Dana Kiley, Gonench Kilich, Shilpa N. Kobren, Isaac S. Kohane, Jennefer N. Kohler, Susan Korrick, Mary Kozuira, Deborah Krakow, Donna M. Krasnewich, Elijah Kravets, Seema R. Lalani, Byron Lam, Christina Lam, Brendan C. Lanpher, Ian R. Lanza, Kimberly LeBlanc, Brendan H. Lee, Roy Levitt, Richard A. Lewis, Pengfei Liu, Xue Zhong Liu, Nicola Longo, Sandra K. Loo, Joseph Loscalzo, Richard L. Maas, Ellen F. Macnamara, Calum A. MacRae, Valerie V. Maduro, Audrey Stephannie Maghiro, Rachel Mahoney, May Christine V. Malicdan, Laura A. Mamounas, Teri A. Manolio, Rong Mao, Kenneth Maravilla, Ronit Marom, Gabor Marth, Beth A. Martin, Martin G. Martin, Julian A. Martínez-Agosto, Shruti Marwaha, Jacob McCauley, Allyn McConkie-Rosell, Alexa T. McCray, Elisabeth McGee, Heather Mefford, J. Lawrence Merritt, Matthew Might, Ghayda Mirzaa, Eva Morava, Paolo Moretti, John Mulvihill, Mariko Nakano-Okuno, Stanley F. Nelson, John H. Newman, Sarah K. Nicholas, Deborah Nickerson, Shirley Nieves-Rodriguez, Donna Novacic, Devin Oglesbee, James P. Orengo, Laura Pace, Stephen Pak, J. Carl Pallais, Christina G. S. Palmer, Jeanette C. Papp, Neil H. Parker, John A. Phillips, Jennifer E. Posey, Lorraine Potocki, Barbara N. Pusey Swerdzewski, Aaron Quinlan, Deepak A. Rao, Anna Raper, Wendy Raskind, Genecee Renteria, Chloe M. Reuter, Lynette Rives, Amy K. Robertson, Lance H. Rodan, Jill A. Rosenfeld, Natalie Rosenwasser, Francis Rossignol, Maura Ruzhnikov, Ralph Sacco, Jacinda B. Sampson, Mario Saporta, Judy Schaechter, Timothy Schedl, Kelly Schoch, Daryl A. Scott, C. Ron Scott, Elaine Seto, Vandana Shashi, Jimann Shin, Edwin K. Silverman, Janet S. Sinsheimer, Kathy Sisco, Edward C. Smith, Kevin S. Smith, Lilianna Solnica-Krezel, Ben Solomon, Rebecca C. Spillmann, Joan M. Stoler, Kathleen Sullivan, Jennifer A. Sullivan, Angela Sun, Shirley Sutton, David A. Sweetser, Virginia Sybert, Holly K. Tabor, Queenie K.-G. Tan, Amelia L. M. Tan, Arjun Tarakad, Mustafa Tekin, Fred Telischi, Willa Thorson, Cynthia J. Tifft, Camilo Toro, Alyssa A. Tran, Rachel A. Ungar, Tiina K. Urv, Adeline Vanderver, Matt Velinder, Dave Viskochil, Tiphanie P. Vogel, Colleen E. Wahl, Melissa Walker, Stephanie Wallace, Nicole M. Walley, Jennifer Wambach, Jijun Wan, Lee-kai Wang, Michael F. Wangler, Patricia A. Ward, Daniel Wegner, Monika Weisz Hubshman, Mark Wener, Tara Wenger, Monte Westerfield, Matthew T. Wheeler, Jordan Whitlock, Lynne A. Wolfe, Kim Worley, Changrui Xiao, Shinya Yamamoto, John Yang, Zhe Zhang, Stephan Zuchner.
Open Research
DATA AVAILABILITY STATEMENT
Both reported variants in ZBTB47 have been deposited in ClinVar: p.(Glu477Lys), accession number: SCV002600228.1; p.(Glu680Gly), accession number: VCV002430193.1.




