Interstitial microdeletions of 3q26.2q26.31 in two patients with neurodevelopmental delay and distinctive features
Funding information: Japan Agency for Medical Research and Development; Ministry of Health, Labour and Welfare; KAKENHI, Grant/Award Number: 21K07873; Japan Society for the Promotion of Science, Initiative on Rare and Undiagnosed Diseases, Grant/Award Number: 20ek0109301
Abstract
Interstitial microdeletions in the long arm of chromosome 3 are rare. In this study, we identified two patients with approximately 5-Mb overlapping deletions in the 3q26.2q26.31 region. Both patients showed neurodevelopmental delays, congenital heart defects, and distinctive facial features. One of them showed growth deficiency and brain abnormalities, as shown on a magnetic resonance imaging scan. Haploinsufficiency of NLGN1 and FNDC3B present in the common deletion region was considered to be responsible for neurodevelopmental delay and the distinctive features, respectively. The possibility of unmasked variants in PLD1 was considered and analyzed, but no possible pathogenic variant was found, and the mechanism of the congenital heart defects observed in the patients is unknown. Because 3q26.2q26.31 deletions are rare, more information is required to establish genotype–phenotype correlations associated with microdeletions in this region.
1 INTRODUCTION
Interstitial microdeletions in the long arm of chromosome 3 are rare. With regards to the 3q26.2q26.31 region, a neonatal case previously reported with a 4.5-Mb deletion in this region showed congenital thrombocytopenia and several distinctive features (Bouman et al., 2016). Since the patient died at 28th day after birth due to respiratory insufficiency, subsequent neurodevelopment of the patient is not known.
Recently, we encountered two Japanese patients with overlapping deletions in this region. The deletions were approximately 5-Mb in size. Both patients showed moderate to severe neurodevelopmental delays, congenital heart defects, and distinctive features, whereas neither showed the hematological abnormalities described in the previous neonatal case. There are some other reports of small deletions in the 3q26.2q26.31 region.
Here, we review the literature and discuss the genotype–phenotype correlation in patients having microdeletions in this region.
2 MATERIALS AND METHODS
This study was conducted and performed in accordance with the Declaration of Helsinki, and the requisite permissions were obtained from the ethical committee of Tokyo Women's Medical University. Peripheral blood samples were drawn from the patients after obtaining written informed consent from their parents. Genomic DNA was extracted from the blood samples using the QIAamp DNA extraction kit (QIAGEN, Hilden, Germany), according to the manufacturer's instructions. Chromosomal microarray analysis (CMA) was performed using the Agilent Microarray 60 K kit (Agilent Technologies), as previously described (Yamamoto et al., 2014). Aberrations in the genomic copy number were visualized using Agilent Genomic Workbench version 7 (Agilent Technologies). In this study, all genomic coordinates were referred as GRCh37/hg19.
In patients having microdeletions in the 3q26.31 region, all coding regions of PLD1 (MIM* 602382) were analyzed by the standard PCR-Sanger sequencing to determine whether they had PLD1 variations in the alternate alleles. The primer sequence is shown in Table S1.
3 RESULTS
3.1 Chromosomal deletions and the genes included in the deletions
Until now, over 3000 samples from undiagnosed patients with neurodevelopmental disorders were analyzed. Among them, overlapping deletions in 3q26.2q26.31 were identified in two different patients (Table 1). Patient 1 showed a 5.36-Mb deletion, described as arr[GRCh37/hg19] 3q26.2q26.31(169,139,502-174,503,530)X1. Patient 2 showed a 5.50-Mb deletion, arr[GRCh37/hg19]3q26.2q26.31(169,272,415-174,775,629)X1. These deletions are depicted on the genome map (Figure 1). The parents of both the patients declined to be genotyped; thus, it was unknown whether the deletions were de novo or inherited.
| Authors published year | Milson et al. (2012) | Nielsen et al. (2012) | Bouman et al. (2016) | Cao et al. (2016) | This study | |||
|---|---|---|---|---|---|---|---|---|
| Patient 1 | Patient 2 | Patient 1 | Patient 2 | |||||
| Age at diagnosis (years) | 6.5 | 1 | newborn | 1.5 | 1.4 | 2.7 | 11.9 | |
| Gender | Female | Female | Male | Female | Male | Male | Female | |
| Growth delay | + | − | NA | − | NA | + | − | |
| Motor delay | + | + | NA | NA | NA | + | + | |
| Intellectual disability | Severe | − | NA | Mild | NA | Severe | Moderate | |
| Behavioral problems | NA | − | NA | NA | NA | + | − | |
| Seizures | + | NA | − | NA | NA | − | − | |
| Distinctive features | ||||||||
| Macrocephaly | − | − | − | + | NA | − | − | |
| Microcephaly | + | + | − | − | NA | − | + | |
| Plagiocephaly | NA | − | − | NA | + | − | − | |
| Large anterior fontanelle | NA | − | − | NA | + | − | − | |
| Prominent forehead | NA | − | − | + | NA | − | − | |
| Midface retrusion | NA | − | − | + | NA | − | − | |
| Flat face | NA | − | − | NA | + | − | − | |
| Cleft palate | NA | − | + | NA | NA | − | − | |
| Low-set ears | NA | − | + | NA | NA | + | + | |
| External ear malformation | NA | − | − | NA | NA | + | + | |
| Proptosis | NA | − | − | NA | + | − | − | |
| Hypertelorism | NA | − | + | NA | + | − | − | |
| Downslanting palpebral fissures | NA | − | − | + | NA | − | − | |
| Blepharophimosis | NA | − | − | NA | NA | + | − | |
| Ptosis | NA | − | − | NA | NA | + | − | |
| Narrow superior/inferior orbital ridges | NA | − | − | NA | + | − | + | |
| Strabisms | NA | − | − | NA | NA | − | + | |
| Upturned nose | NA | − | − | + | NA | − | + | |
| Shallow nasal bridge | NA | − | − | NA | + | − | − | |
| Broad nasal bridge | NA | − | + | NA | NA | − | + | |
| Underdeveloped ala nasi | NA | − | − | NA | NA | + | − | |
| Anteverted nostrils | NA | − | − | NA | + | − | − | |
| Short nose | NA | − | − | NA | + | − | − | |
| Thin upper lip | NA | − | + | NA | NA | − | − | |
| Brain anomaly | Diffuse and non-specific increased T2 signal in white matter | NA | Two small cysts of the choroid plexus | Hydrocephaly | NA | Nonspecific, symmetrical high-intensity areas in deep white matter | − | |
| Congenital heart defects | NA | NA | Ventricular septal defects | NA | NA | Mild pulmonary valve stenosis | Tricuspid valve dysplasia and patent ductus arteriosus | |
| Extremities | ||||||||
| Abnormal muscle tone | + | NA | NA | NA | NA | − | − | |
| Joint laxity | NA | NA | NA | + | NA | − | − | |
| Flat feet | NA | − | − | + | NA | − | + | |
| The other abnormalities | Prominent digit pads of fingers and toes | Clubbing of the fingernails | − | NA | NA | Palmar crease | Short fourth metacarpal short ulna | |
| Other congenital abnormalities | NA | Thrombocytopenia Subsequent aplastic anemia |
Thrombocytopenia | NA | NA | Hypertrophic pyloric stenosis | Hypertrichosis | |
| Affected chromosomal band | 3q26.31q26.32 | 3q26.2 | 3q26.2q26.31 | 3q26.31 | 3q26.31 | 3q26.2q26.31 | 3q26.2q26.31 | |
| Genomic positiona | ||||||||
| Start | 173,855,599 | 168,467,339 | 168,980,281 | 171,677,940 | 171,622,716 | 169,139,502 | 169,272,415 | |
| End | 176,104,161 | 169,218,686 | 173,518,426 | 172,191,359 | 171,968,102 | 174,503,530 | 174,775,629 | |
| Size of deletion (Mb) | 2.249 | 0.751 | 4.54 | 0.513 | 0.345 | 5.364 | 5.503 | |
| The number of genes involved | 2 | 2 | 27 | 2 | 1 | 27 | 28 | |
| FNDC3B included | − | − | + | + | + | + | + | |
| NLGN1 included | + | − | + | − | − | + | + | |
- a Genomic positions are referred to GRCh37/hg19.
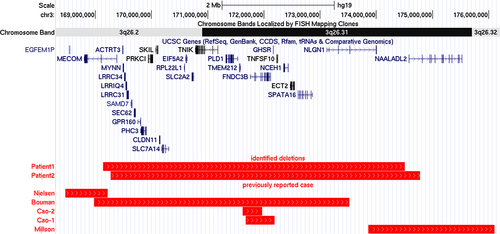
A total of 28 coding genes were included in the deletion region. According to the Online Mendelian Inheritance in Man® (OMIM®; https://www.omim.org/), 10 are associated with human phenotypes (Table S1).
3.2 Clinical reports
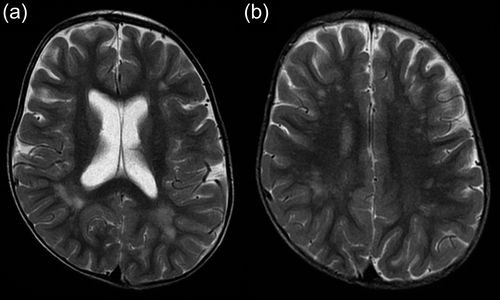
Patient 1 was a 12-year-old boy, the first child of a healthy couple in Japan. This patient was born at 33 weeks and 6 days of gestation by caesarean section for fetal distress without neonatal asphyxia (the Apgar scores were not available). His birth weight was 1870 g (−0.7 standard deviation [SD]), height was 39 cm (−1.9 SD), and occipitofrontal circumference (OFC) was 33 cm (+1.4 SD). At 2 years of age, his height was 77 cm (−4.1 SD), weight was 7.9 kg (−3.7 SD), and OFC was 49 cm (0 SD), indicating a growth delay from infancy. At present, his height is 130 cm (−2.3 SD), weight is 25.8 kg, and his Rohrer's index of 116.8 is within normal limits. To address the issue of heart murmur during infancy, echocardiography was performed, which detected a mild pulmonary valve stenosis. Brain magnetic resonance imaging (MRI) was performed at 2.5 years of age (Figure 2). There were no findings such as brain atrophy or hydrocephalus. Bilateral spotty areas of T2-high intensity were diffusely observed in the deep white matter and more prominent in the parietal and occipital lobes. This finding was considered as non-specific for neurological disorders.
His gross motor development was delayed; he achieved head control at 18 months, sat without assistance at 24 months, and did not take his first steps until 32 months. Currently, he can walk unsupported till a certain distance but tumbles down easily. Language development is delayed, and he is still nonverbal. He has maladaptive behaviors including self-injury, scratching himself, and throwing temper tantrums. He shows distinctive clinical features, including low-set ears, underdeveloped alae nasi, blepharophimosis, ptosis of the left eye, and single transverse palmar crease on the left hand. Conventional GTG-banding detected an interstitial deletion in 3q, described as 46,XY,del(3)(q26.2q26.3).
Patient 2 was a 12-year-old Japanese girl, who presented with neurodevelopmental delay and distinctive features. She was born at 37 weeks of gestation, with a weight of 2178 g (−1.1 SD), a height of 44.2 cm (−1.3 SD), and an OFC of 29.5 cm (−2.1 SD). Echocardiography performed during the neonatal period revealed tricuspid valve dysplasia and patent ductus arteriosus (PDA). Thus, a surgical closure was performed for PDA in infancy. Brain MRI conducted at 1 year of age did not reveal any abnormalities.
Her motor developmental milestones were delayed; head control at 4 months, rolling at 12 months, sitting at 24 months, crawling at 32 months, standing with support at 36 months, and walking at 48 months. Language development was also delayed; a single word was present at 36 months and three-word sentence at 7½ years.
At present, her physical growth is within the normal limits; her height is 140.9 cm (−1.3 SD), and her weight is 35.6 kg (−0.8 SD). Her development is delayed. Currently, she attends a special educational program. Although her activities of daily living were not so impaired and she did not show any impaired communication skills or behavioral abnormalities, the Tanaka-Binet intelligence scale score was 48 at 12 years of age, indicating moderate intellectual disability. Conventional chromosomal GTG-banding revealed a normal female karyotype of 46,XX.
She shows distinctive features, including low-set ears, narrow superior orbital ridges, strabismus, upturned nose, broad nasal bridge, flat feet, short fourth metacarpal, short ulna, and hypertrichosis.
There were no findings of abnormal supination and pronation in both hands of Patients 1 and 2.
3.3 PLD1 analysis
All coding regions of PLD1 were analyzed and a missense variant, NM_002662.5(PLD1):c.2576G>A (chr3:171360647-C-T[GRCh37/hg19], p.Gly859Gln, rs186118100), was detected in Patient 2. The global allele frequency (0.0001785) was low in gnomAD v2.1.1 (https://gnomad.broadinstitute.org/), whereas it was relatively high in Japanese population as 0.021 according to the Human Genetic Variation Database (https://www.hgvd.genome.med.kyoto-u.ac.jp/index.html) (Higasa et al., 2016).
4 DISCUSSUON
In this study, we identified two overlapping interstitial microdeletions spanning the 3q26.31-q26.32 region in two different patients (Figure 1). Both patients showed distinctive facial features, neurodevelopmental delay, and congenital heart defects. Especially, one of the patients (Patient 1) showed severe psychomotor developmental delay accompanied with non-specific brain MRI findings. The congenital heart anomalies observed in both patients were mainly on their right side. Although congenital thrombocytopenia was described in the previously reported neonatal patient (Bouman et al., 2016), hematological abnormalities were not observed in our patients.
Interstitial deletions in this region are rare, but a few clinical reports have been published previously (Figure 1). Cao et al. reported two patients with small deletions in a narrow region associated with craniofacial characteristics (Cao et al., 2016). Because the fibronectin type III domain containing the 3B gene (FNDC3B; MIM *611909) was located in the shortest region of overlap, this gene was considered a possible candidate responsible for the craniofacial features of the patients (Cao et al., 2016). FNDC3B was also deleted in the patient reported by Bouman et al. (2016), as well as in the patients described in this study. Referring to their clinical features (Table 1), it was inferred that all patients having deletions including FNDC3B show distinctive facial features. FNDC3B, the human ortholog of mouse fad104, has been suspected to have a potential role in craniofacial development, based on the research findings in a mouse model. Kishimoto et al. reported that fad104-deficient mice had several cranial and skeletal malformations (Kishimoto et al., 2010). Therefore, the distinctive features of the patients were considered to be the consequence of FNDC3B haploinsufficiency.
The neonatal patient reported by Bouman et al. had a 4.5-Mb deletion in 3q26.2q26.31 (Bouman et al., 2016). The patient exhibited congenital thrombocytopenia and distinctive features. The proximal region of the deletion overlapped with that reported by Nielsen et al. (2012). Because the patients reported by both Bouman et al. and Nielsen et al. showed thrombocytopenia, the MDS1 and EVI1 complex locus (MECOM; MIM *165215) located in the shortest region of overlap was considered to be the gene responsible for the hematological phenotype. In the Online Mendelian Inheritance in Man® (OMIM, https://www.omim.org/), the phenotype associated with MECOM is radioulnar synostosis with amegakaryocytic thrombocytopenia 2 (MIM #616738). However, the two patients reported showed neither feature. This could be due to the clinical heterogeneity of deletion in MECOM, as reported by Germeshausen et al. (2018).
Distal regions of the shortest region of overlap in these two patients overlapped with the deletion region identified in a previously reported patient with microcephaly, seizure disorder, and severe ID (Millson et al., 2012). In the shortest overlapping region, only two genes, NLGN1 (MIM *600568) and NAALADL2 (MIM* 608806), were involved. Among them, NLGN1 (Neuroligin 1), a member of the NLGN protein family, is a major component of excitatory glutamatergic synapses, and several missense variants have been reported to be associated with ASD (Nakanishi et al., 2017). Another study suggested the relationship between NLGN1 and ID/ASD (Tian et al., 2021). However, the precise clinical burden of the variations in NLGN1, especially owing to deletions, is still unclear because of its rarity. As is the case with individuals having missense variations in NLGN1, it seems that individuals with NLGN1 deletions do not always develop neurodevelopmental disorders. According to the case report from Millson et al. (2012), the patient's unaffected mother harbored the same deletion as the patient. This is acceptable because neurodevelopmental disorders, including ASD, are multifactorial, and the cumulative effect of various risk factors leads to the onset of diseases (Leppa et al., 2016). Accumulation of more clinical data would be necessary to establish the clinical burden of NLGN1 deletions.
The patient reported by Millson et al. showed severe ID and abnormal intensity of the diffuse non-specific deep white matter, as detected on the brain MRI scan, similar to those in Patient 1 in this study (Millson et al., 2012). Millson et al. suspected that the NLGN1 deletion contributed to the patient's infantile spasms and caused severe neurological phenotypes (Millson et al., 2012), whereas Patient 1 in this study did not experience spasms or seizures. These findings indicate the possible clinical heterogeneity of NLGN1 haploinsufficiency.
In this study, the 3q26.31q26.32 deletion region contained CLDN11 (MIM *601326). CLDN11 (Claudin-11), a member of the claudin tight junction family, is an important element of the myelin sheath and has been reported to act as a barrier to diffusion (Denninger et al., 2015). Riedhammer et al. reported three patients with hypomyelinating leukodystrophy who harbored a common stop-loss pathogenic variant, and these patients presented with spasms, neurodevelopmental delay, and ocular abnormalities (Riedhammer et al., 2021). The identified variant was suspected to have a dominant-negative effect. Therefore, haploinsufficiency of CLDN11 would not contribute to any abnormality. Although the patient with the 3q26.2q26.31 deletion reported by Bouman et al. harbored both NLGN1 and CLDN11 in the deleted region (Bouman et al., 2016), this patient had been reported to have died during the neonatal period owing to respiratory insufficiency; thus, information regarding neurodevelopment was unavailable.
Both the patients in this study had congenital heart defects (CHDs) mainly on the right side. Previously, recessive variants of PLD1 (phospholipase D1; MIM* 602382) were reported to be associated with right-sided congenital cardiac valve defects (Ta-Shma et al., 2017), and recently, Lahrouchi et al. confirmed that biallelic loss-of-function (LOF) variants of PLD1 caused the right-sided congenital cardiac valve defects (Lahrouchi et al., 2021). PLD1 is a signal transduction enzyme that hydrolyzes the membrane lipid phosphatidylcholine to generate a second messenger. It is reported that the patients carrying biallelic LOF variants had reduced enzymatic activity (lower than 25%) compared with that of individuals having wild-type alleles (Lahrouchi et al., 2021). It was also noted that the LOF of PLD1 leads to a suppression of TGF-β and sonic hedgehog signaling, which might induce arterial pole defects in the developing heart. Furthermore, they reported that none of the patients with biallelic variations in PLD1 had dysmorphic features, ID, or neurodevelopmental delays, inferring that these variations were predominantly associated with isolated cardiac diseases. Because the patient reported by Bouman et al. also showed congenital heart defect (Bouman et al., 2016), we considered contribution of PLD1 to the congenital heart defect observed in the patients in this study and the possibility of an unmasked PLD1 mutation in the remaining allele was suspected. Although this study identified a missense variant in Patient 2, general frequency of this variant in Japanese individuals was high and there was no positive information to support that this variant as a pathogenic to congenital heart defect in Patient 2. The precise clinical burden of the deletions including PLD1 remains unclear because of the lack of evidence from the clinical literature; however, it cannot be ruled out that PLD1 haploinsufficiency may be associated with the development of CHD.
In this study, two additional patients with 3q26.2q26.31 deletions were identified. Some genes were considered to contribute to some clinical features, but more information would be required to establish genotype–phenotype correlation in patients having microdeletions in this region.
AUTHOR CONTRIBUTIONS
Toshiyuki Yamamoto designed this study. Takeaki Tamura involved in organizing this study and drafting the manuscript. Keiko Shimojima Yamamoto, Takashi Shiihara, Satoru Sakazume, and Nobuhiko Okamoto have contributed to acquisition of data. Hiroshi Yagasaki, Ichiro Morioka, and Hitoshi Kanno critically reviewed the manuscript. All authors contributed to the analysis and interpretation of data. All authors agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
ACKNOWLEDGMENTS
We would like to express our gratitude to the patients and their families for their cooperation.
FUNDING INFORMATION
This study was supported by KAKENHI (Grant Numbers 21K07873) from the Japan Society for the Promotion of Science, Initiative on Rare and Undiagnosed Diseases (grant number 20ek0109301) from the Japan Agency for Medical Research and Development (AMED), and a grant from the Ministry of Health, Labor, and Welfare Japan.
CONFLICT OF INTEREST
The authors declare no conflicts of interest or competing interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.




