Copy number variants suggest different molecular pathways for the pathogenesis of bladder exstrophy
Funding information: Swedish Research Council, Grant/Award Number: K2012-64X-14506-10-5 2016–01642; Foundation Frimurare Barnhuset Stockholm; Stockholm City Council; Karolinska Institutet; the Swedish Brain Foundation; the Harald and Greta Janssons Foundation; Erik Rönnbergs Foundation
Abstract
Bladder exstrophy is a rare congenital malformation leaving the urinary bladder open in the midline of the abdomen at birth. There is a clear genetic background with chromosome aberrations, but so far, no consistent findings apart from 22q11-duplications detected in about 2%–3% of all patients. Some genes are implicated like the LZTR1, ISL1, CELSR3, and the WNT3 genes, but most are not explained molecularly. We have performed chromosomal microarray analysis on a cohort of 140 persons born with bladder exstrophy to look for submicroscopic chromosomal deletions and duplications. Pathogenic or possibly pathogenic microdeletions or duplications were found in 16 patients (11.4%) and further 9 with unknown significance. Two findings were in regions linked to known syndromes, two findings involved the same gene (MCC), and all other findings were unique. A closer analysis suggests a few gene networks that are involved in the pathogenesis of bladder exstrophy; the WNT-signaling pathway, the chromosome 22q11 region, the RIT2 and POU families, and involvement of the Golgi apparatus. Bladder exstrophy is a rare malformation and is reported to be associated with several chromosome aberrations. Our data suggest involvement of some specific molecular pathways.
1 INTRODUCTION
Bladder exstrophy is a congenital malformation and part of a clinical spectrum of the bladder exstrophy-epispadias complex (BEEC, Online Mendelian Inheritance in Man [OMIM] 600057). The phenotypic severity varies from isolated epispadias, to the most severe and rare form, cloaca exstrophy, with the most common variant classic bladder exstrophy (CBE) of medium severity. The incidence of CBE in Sweden is 1 in 30,000 live births (Reinfeldt Engberg et al., 2016). Most cases are sporadic, however, an increased risk in siblings has been noted. Shapiro et al. (1984) reported a recurrence risk for a sibling of 1/70, equivalent to a 400 times increased risk compared with the general population. About 3% of all cases are classified as familial with at least two affected persons in the same family (Reutter et al., 2003). In addition, the pairwise concordance rates in monozygotic as compared with dizygotic BEEC twin pairs are 45% and 6%, respectively (Reutter et al., 2007). Finally, several chromosomal aberrations have been published in association with CBE (Ludwig et al., 2005; von Lowtzow et al., 2016). These combined data strongly suggest an underlying genetic component involving several different genes.
The only recurrent genetic finding so far is the 22q11-duplication encompassing ~3 Mbp and including 50 genes (Draaken et al., 2010, 2014; Lundin et al., 2010, 2019; Pierquin & Uwineza, 2012). The 22q11 duplication syndrome was initially described in 2008 and was mainly associated with cognitive deficits and dysmorphic facial features. The syndrome has a large clinical variability and reduced penetrance, since healthy and near-healthy carriers are common (Courtens et al., 2008). Data from different bladder exstrophy cohorts have shown 22q11 duplications in 2.5% of all patients, thus giving an OR of around 30 for being born with bladder exstrophy in carriers. The exact molecular mechanism for how 22q11duplication increases the risk is still unclear. The candidate region was diminished after new findings of smaller regions of overlap and together with expression pattern the main candidate genes are the CRKL, THAP7, and LZTR1 genes (Draaken et al., 2014). In addition, our group detected one novel mutation in the LZTR1 gene located in this region in a patient with CBE and could show that the mutation has functional effect in the Golgi apparatus (Lundin et al., 2019).
Some specific genes have been reported to be associated with BEEC. This includes the ISL1-gene on chromosome 5q11.1, which was identified in a GWAS and with a few mutational findings (Arkani et al., 2018; Draaken et al., 2015). Through whole exome sequencing in CBE a de novo mutation was detected in the WNT3-gene, which also caused defect cloaca development in zebrafish (Baranowska Körberg et al., 2015). Further, three missense mutations have been identified in the SLC20A1-gene on chromosome 2q13, detected in cloacal exstrophy and CBE (Reutter et al., 2016; Rieke et al., 2020). Also, compound heterozygous mutations in the CELSR3-gene, on chromosome 3p21.31, were detected in a child with cloacal exstrophy and other malformations (Reutter et al., 2016).
These findings prompted us to perform a genome-wide search for deletions and duplications using chromosomal microarray analysis (CMA). The aim of this study was to further evaluate copy number variants (CNVs), in a cohort of 140 well-characterized Swedish BEEC patients to identify additional genetic events associated with BEEC.
2 MATERIAL AND METHODS
2.1 Editorial policies and ethical considerations
The study was approved by the Swedish Ethics Review Authority and conforms to the Declaration of Helsinki standards. All patients or parents gave their informed consent prior to inclusion in the study.
2.2 Patients
All BEEC patients, adults and children, were recruited from the Pediatric Surgery Departments in Stockholm, Göteborg, Uppsala, and Lund, Sweden. Six patients with previously published chromosome aberrations were excluded from the study (Lundin et al., 2010, 2019; Soderhall et al., 2014). Blood or excess bladder or skin tissue obtained during surgery was collected for DNA isolation and molecular genetic analysis. When possible, parental DNA was also collected.
2.3 Chromosomal microarray
A 4 × 180 K custom oligonucleotide microarray with whole-genome coverage and a median probe spacing of ~18 kb was used (AMADID:031035, Oxford Gene Technology, Begbroke, Oxfordshire, UK). This array design is used as a routine diagnostic tool at the Department of Clinical Genetics, Karolinska University Hospital, Stockholm, Sweden.
The control DNA used for the microarray experiment consisted of a mix of sex-matched DNA from several healthy individuals pooled together (Promega, Madison, Wisconsin). Sample labelling (CGH labelling kit for oligo arrays, Enzo Life Sciences, Farmingdale, New York), hybridization, and slide washing (Oligo aCGH/ChIP-on-Chip Wash Buffer Kit, Agilent Technologies, Wilmington, Delaware) were performed according to the manufacturers' recommendations. Slides were scanned using the Agilent Microarray Scanner (G2505C, Agilent Technologies, USA) with 3 μm resolution. Raw data were normalized using Feature Extraction Software v10.7.3.1 (Agilent Technologies, Santa Clara, California), and log2 ratios were calculated by dividing the normalized intensity in the sample by the mean intensity across the reference sample. The log2 ratios were plotted and segmented by circular binary segmentation in the CytoSure Interpret software v4.10 (Oxford Gene Technology, Oxfordshire, UK). Oligonucleotide probe positions were annotated according to the human genome assembly hg19 (Genome Reference Consortium Human Build 37). For the 4 × 180 K microarray, three consecutive aberrant probes with a log2 ratio cutoff of − 0.65 for deletions and 0.35 for duplications were called, giving a practical lower resolution of about 50 kb. The clinical relevance of all CNVs was classified into five categories: benign, likely benign, variant of uncertain significance, likely pathogenic, and pathogenic, according to the American College of Medical Genetics and Genomics guidelines (Kearney et al., 2011). Classification was based upon the size of aberration, gene content, inheritance, and available information in medical literature and different databases: The Database of Genomic Variants (DGV, 2021), The Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER, 2021), The Online Mendelian Inheritance in Man (OMIM, 2021), and an in-house database with variants from >10,000 analyzed clinical patients referred to the Department of Clinical Genetics for CMA. Further information on genes that were involved in deletions or duplications was collected from OMIM, GeneCards—The Human Gene Database (2021), and MalaCards (www.genecards.com; Stelzer et al., 2016).
2.4 Fetal data for comparison
RNA expression data obtained from 17 samples of embryonic and fetal bladders (fetal Weeks 5–10) were used for evaluating expression of individual genes in deleted or duplicated CNVs. Data are available publicly at https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-6592/protocols/.
3 RESULTS
3.1 Clinical description
We analyzed DNA from 140 individuals with BEEC. Of these, 114 were born with CBE, 14 with epispadias and 12 with cloacal exstrophy and none was a familial finding. Using CMA, we identified 16 microdeletions or duplications (11.4%) that were classified as findings with possible associations to the pathophysiology of BEEC (Table 1). Two were rearrangements in regions previously associated with known syndromes, and 14 were evaluated as likely pathogenic or possibly pathogenic after a standard clinical evaluation. Of these 16, 12 had CBE, one was born with epispadias, two had cloacal exstrophy and in one male the bladder was extremely small, bordering on bladder agenesis. One patient also had cleft lip and palate and one had duodenal atresia. Nine additional samples exhibited CNVs that in a clinical setting would be regarded as of unknown significance (Table 2). Altogether, two patients had two CMA findings (Tables 1 and 2).
| Locus | Position (hg38) | Size | Dup/Del | Phenotype | Sex | Inheritance | Ref Seq genes | Fetal expr | Other dba |
|---|---|---|---|---|---|---|---|---|---|
| Two findings in syndromic regions | |||||||||
| 16p11.2 | chr16:29645396–30168276 | 523 kb | Del | CBE, ASD | M | NA | QPRT, SPN, SEZ6L2, PAGR1, MVP, ZG16, KIF22, FAM57B, HIRIP3, TAOK2, GDPD3, MAZ, PRRT2, CDIPT, KCTD13, ASPHD, THEM219, ALDOA, DOC2A, INO80E, C16orf92, HIRIP3, TBX6, PPP4C, YPEL3, MAPK3 | High expression in bold | Known syndrome |
| Xq28 | chrX:154822249–155197455 | 375 kb | Dup | CBE and duodenal stenosis | F | NA | SMIM9, F8, FUNDC2, CMC4, MTCP1, BRCC3, VBP1 | Yes, all | Known syndrome |
| Possibly pathogenic findings | |||||||||
| 1p36.11 | chr1:26002697–26255559 | 253 kb | Del | CBE | M | Mat | EXTL1, SLC0A2, TRIM63, PDIK1L, FAM110D, ZNF593, CNKSR1, CATSPER4, CEP85 | Yes, all | Novel |
| 3q26.1 | chr3:160985377–164524799 | 3.54 Mb | Dup | Epispadias | M | PPM1L, B3GALNT1, NMD3, SPTSSB | Yes | A few similar reported | |
| 5q22.2 | chr5:113180369–113318123 | 138 kb | Del | CBE | M | MCC | Yes | Only larger del reported | |
| 5q22.2 | chr5:113001507–113151488 | 150 kb | Dup | Cloacal exstropy and cleft palate | F | Pat | MCC | Yes | Only larger dup reported |
| 6q23.2 | chr6:133892118–133892292 | 175 kb | Dup | CBE | F | TCF21 | Yes | Novel | |
| 8q22.2 | chr8:98278761–98644873 | 366 kb | Del | CBE | F | Pat | NIPAL2, KCNS2, STK3 | Yes | One smaller reported |
| 8q22.2 | chr8:99529972–99624795 | 95 kb | Del | CBE | M | Mat | VPS13B | Yes | Similar reportedb |
| 9p13.2 | chr9:36363506–36944808 | 581 kb | Dup | CBE | F | Mat | RNF38, MELK, PAX5 | Yes | Three similar reported |
| 9p22.2 | chr9:18351191–18668331 | 317 kb | Dup | CBE | F | Pat | ADAMTSL1 | Yes | A few reported |
| 10p12.2 | chr10:24091999–24375694 | 284 kb | Dup | Bladder agenesis | M | Pat | KIAA1217 | Yes | A few reported |
| 16q24.3 | chr16:89752071–89842991 | 91 kb | Del | CBE | M | Mat | FANCA, SPIRE2 | Yes | Reported earlier |
| 18q12.3 | chr18:43052727–43463356 | 411 kb | Del | CBE | M | Pat | RIT2, SYT4 | Yes | Novel |
| 21q22.12 | chr21:34671646–34731540 | 60 kb | Dup | CBE | Fc | Mat | CLIC6 | Yes | Novel |
| Xq23 | chrX:115139141–115189073 | 50 kb | Del | CBE | M | Mat | LRCH2 | Yes | Novel |
- Note: Genes in bold have a high expression in fetal bladder, gestational weeks 5–10 (Expression Atlas - E-MTAB-6592).
- Abbreviations: ASD, autism spectrum disorder; BEEC, bladder exstrophy-epispadias complex; CBE, classic bladder exstrophy; DECIPHER, The Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources; DGV, The Database of Genomic Variants; OMIM, The Online Mendelian Inheritance in Man.
- a Searches for similar variants have been performed in DGV, DECIPHER, OMIM and our in-house database with variants from >10,000 analyzed clinical cases referred for CMA as reported in January 2022.
- b Two cases with urogenital malformations among those reported.
- c Same individual with two findings, Tables 1 and 2.
| Locus | Position (hg38) | Size (kb) | Dup/Del | Phenotype | Sex | Inheritance | Ref Seq genes | Fetal expr | Detected in other dba |
|---|---|---|---|---|---|---|---|---|---|
| 2q35 | chr2:220073652-220182172 | 109 kb | Del | Cloacal exstrophy | M | ZFAND2B, ABCB6, ATG9A, ANKZF1, GLB1L, STK16, TUBA4A, DNAJB2, and PTPRN | Yes | Less than 10 similar | |
| 3p25.3 | chr3:10259315–10285321 | 25 | Del | CBE | Mb | TATDN2 | Yes | Three larger reported | |
| 3q21.1 | chr3:123180879–123276259 | 95 | Del | CBE | Fc | SEC22A | Yes | Less than 10 similar | |
| 4p16.3 | chr4:342319–426951 | 85 | Del | CBE | F | ZNF141 | Yes | Similar reported | |
| 7p22.2 | chr7:3406217–3642243 | 236 | Dup | CBE | M | Mat | SDK1 | Yes | Several reported |
| 8p23.1 | chr8:11809436–11875305 | 66 | Del | CBE | F | Pat | FDFT1 and CTSB | Yes | Around 10 reported |
| 8p22 | chr8:15218133–15334651 | 117 | Del | Cloacal exstrophy | M | SGCZ | Yes | Several reported in the region | |
| 9q31.1 | chr9:99907569–99959918 | 52 | Dup | CBE | F | STX17 | Yes | Only two reported | |
| 19q13.2 | chr19:40819832–41120423 | 301 | Dup | CBE | Mb | Pat | BCKDHA, THEM91, ERICH4, DMAC2, B9D2, TGFB1, EXOSC5, B3GNT8, TPM3P5, and CEACAM21 | Yes, apart from ERICH4 | Around five reported |
- Note: Genes in bold have an established expression in fetal bladder gestational week 5–10 (Expression Atlas - E-MTAB-6592).
- Abbreviations: BEEC, bladder exstrophy-epispadias complex; DECIPHER, The Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources; DGV, The Database of Genomic Variants; OMIM, The Online Mendelian Inheritance in Man.
- a Searches for similar variants have been performed in DGV, DECIPHER, OMIM, and our in-house database with variants from >10,000 analyzed clinical cases referred for CMA as reported in January 2022.
- b Same individual with two findings, Table 2.
- c Same individual with two findings, Tables 1 and 2.
3.2 CNV findings
Data on microarray findings are listed in Tables 1 and 2, including chromosomal region, size of the deletion/duplication, phenotype, sex, parental status when available, genes within the affected region, and whether the genes within the region are expressed in human fetal urinary bladder. Information on genes was collected from OMIM and GeneCards as described above. The aberrations detected in the study were not recurrent apart from 5q22.2 where two cases had aberrations (one deletion and one duplication) involving different parts of the same gene, the MCC Regulator of Wnt Signaling pathway gene, MCC.
3.3 Two findings in syndromic regions
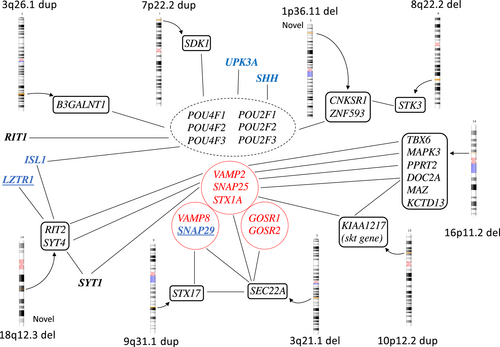
One DNA sample showed a “16p11.2 deletion” (523 kb), encompassing 24 known genes and covering almost the whole region of the classic 16p11.2 deletion syndrome (Table 1, Figures 1 and 2). This contiguous gene syndrome typically covers 593 kb and is often associated with hypospadias and other forms of urogenital malformations (Nik-Zainal et al., 2011; Sampson et al., 2010; Tannour-Louet et al., 2010). Among those with urogenital malformations, the smallest common region of 217 kb contains the MYC-Associated Zinc Finger Protein, MAZ, that is a dosage-sensitive regulator in urogenital development and crucial for normal bladder development (Haller et al., 2018). Our case with a 523 kb deletion and a neuropsychiatric disorder is the first reported with bladder exstrophy. The mechanism could be associated with WNT-gene expression. The deleted region also contains two additional genes that are potentially interesting. The T-box transcription factor 6 gene (TBX6), is involved in the fetal development in early mesoderm and described as the driver for renal and urinary tract malformations in the 16p11-deletion syndrome (Verbitsky et al., 2019). The Potassium Channel Tetramerization Domain-containing protein 13 (KCTD13) gene is expressed in fetal bladder and may also be a candidate for urogenital malformations. There are also other genes highly expressed in fetal bladder in this region, marked in bold in table 1. The Double C2-like domain-containing protein alpha, DOC2A, is involved in fusion between membranes. It is localized mainly in the nucleus and is associated with several Golgi proteins like SNAP25, SNAP29 (in the 22q11 duplication region), and STX1B (Sato et al., 2010, GeneCards).
One female with CBE in combination with duodenal stenosis carried a 375 kb duplication on chromosome Xq28 containing only a few genes: Small integral membrane protein 9 (SMIM9), Coagulation factor VIII (F8), FUN14 Domain Containing protein 2 (FUNDC2), and C-X9-C Motif containing 4 (CMC4), Mature T-cell proliferation 1 (MTCP1), and the BRCA1/BRCA2-containing complex, subunit 3 (BRCC3). In most patients, the Xq28 duplication syndrome covers a larger region and includes the gene MECP2, that is regarded as causative of the intellectual disability and cognitive disturbances. The syndrome has been described as associated with bladder dysfunction (Clayton-Smith et al., 2009).
3.4 CNV-findings regarded as possibly pathogenic
One male with a CBE carried a maternally inherited 1p36.11 deletion (253 kb), involving nine genes. This deletion is not described in any public databases. Three of the genes are active in skeletal or muscle tissue that could be involved in the pathogenesis in BEEC, namely, the Exostosin-like glycosyltransferase 1 (EXTL1) the Tripartite Motif-Containing protein 63 (TRIM63), and the Centrosomal protein, 85-KD, CEP85 genes. The gene products of EXTL1 and CEP85 are mostly localized in the Golgi apparatus. The Zinc-Finger Protein 593 (ZNF593) gene is a negative regulator of OCT2 (the POU2F2 gene; Terunuma et al., 1997).
One larger 3q26.1-duplication (3.54 Mb), containing four genes, was identified in a male with epispadias. A few similar duplications are reported in databases. All the duplicated genes are expressed in fetal urinary bladder, supporting the hypothesis that one or more of the genes contributed to the malformation. The NMD3 ribosome export adaptor (NMD3) is located in the nucleus. The Serine Palmitoyltransferase, small subunit B (SPTSSB) gene is associated with Golgi-proteins. The Protein Phosphatase, magnesium or manganese requiring protein (PPM1L), is expressed in muscle. It is involved in initiating an apoptosis cascade and associated with Golgi function. The beta-1,3-N-acetylgalactosaminyltransferase 1 gene (B3GALNT1) is highly expressed in the Golgi apparatus, and is active in embryonal ectoderm, in urinary bladder and is associated with the POU2F3 gene.
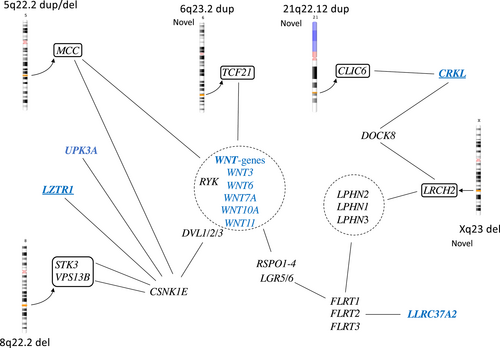
On chromosome 5q22.2, aberrations were detected in two patients, one with CBE and one with cloacal exstrophy. Both aberrations, one paternally inherited 150 kb duplication and one 138 kb deletion of unknown inheritance, were in the same gene but not covering overlapping regions. The gene MCC Regulator of Wnt Signaling pathway, MCC, is a tumor suppressor gene and somatic mutations have been detected in colorectal cancer. The gene is expressed in urinary bladder and this region is also described in bladder cancer progression (Majewski et al., 2008; von Knobloch et al., 2000).
One interesting duplication finding on chromosome 6 (6q23.2) contains the 3′ end of the gene Transcription Factor 21 (TCF21), that is active in the embryological mesoderm surrounding the genitourinary system and a prognostic marker in bladder cancer (Lotfi et al., 2021). This duplication has not been described previously. The gene is associated with both canonical and non-canonical WNT-pathways as well as with the ALDOA gene, in the 16p11.2 deletion, described above.
Two non-overlapping 8q22.2 deletions were found in a male and a female with CBE. One was a unique 366 kb deletion containing three genes. First, the NIPA Like Domain Containing 2, NIPAL2, which codes for a membrane protein, earlier named SLC57A4, that has been associated with ichtyosis. Second, the Potassium Channel, Voltage-Gated, Delayed-Rectifier7, Subfamily S, Member 2, KCNS2, which codes for a membrane protein only expressed in brain tissue. Last, the Serine/Threonine Protein Kinase 3 (STK3), a growth suppressor that is expressed during human fetal development, also in urinary bladder, and especially important for vasculogenesis. STK3, together with its homolog STK4, is part of the Hippo pathway that is important during development for growth and apoptosis. The STK3/4 genes are also associated with the CNKSR1 gene in the chromosome 1p36.11-deletion. All three deleted genes are expressed in fetal bladder, although KCNS2 to a small degree. This deletion was also found in the father.
The second 8q22.2 deletion was 95 kb, inherited from the mother and involved only one single gene, the Vacuolar Protein Sorting 13 homolog B, VPS13B. It is expressed in most tissues including fetal urinary bladder, and mutations cause the autosomal recessive Cohen syndrome with intellectual disability, microcephaly, and epilepsy, as well as other symptoms. VPS13B is a transmembrane protein important in vesicle-mediated transport (Rodrigues et al., 2018), maybe post-Golgi. In vitro knock-down has shown alterations in the Golgi, lysosome and endosome morphology (Duplomb et al., 2014). Gains of the 8q22 region have been described in bladder cancer (Richter et al., 1999). This patient also carried a previously reported ISL1-mutation (Arkani et al., 2018). Similar deletions have been reported in-house in two patients with gastrointestinal and kidney malformations.
Three genes reside within the 9p13.2 duplication (581 kb) that was found in a female with CBE and her mother. The Ring Finger Protein 38, RNF38, is expressed in different tissues and active in development and oncogenesis as well as regulation of p53 (Sheren & Kassenbrock, 2013). The Maternal Embryonic Leucine Zipper Kinase, MELK, codes for a protein kinase that is important in early embryonic development for example in skeletal muscle and during oncogenesis (Jiang & Zhang, 2013). Both these genes are highly expressed in fetal bladder. The Paired Box Protein 5, PAX5, is crucial for B-cell differentiation. Similar aberrations have been reported in a few cases in public databases.
The 9p22.2p22.1 duplication (317 kb), found in a female with CBE only encompasses the gene Adamts-like Protein 1, ADAMTSL1, which is initially expressed at a low level in fetal bladder, increasing over time during bladder development. The glycoprotein is found extracellularly and in the endoplasmatic reticulum. The gene is expressed in skeletal muscle and may be involved in a midline orientation network (Seetharaman et al., 2011). Interestingly, CBE has also been described in a case of Opitz syndrome, a midline disorder (Jacobson et al., 1998). A few cases have been reported in public databases. The duplication in our patient was found paternally inherited.
The gene KIAA1217 (Sickle tail, mouse, homolog of, SKT) is duplicated in the chromosomal region 10p12.2p12 (284 kb). Such a duplication was not found in the in-house database, but a few in public databases. The duplication was paternally inherited. The gene is expressed in many tissues, especially in intervertebral discs. KIAA1217 corresponds to the sickle tail (skt) gene in mice and is expressed also in the cloacal plate during fetal development (Semba et al., 2006; Suda et al., 2011). This gene is of special interest due to association and network with the paralogue gene, SRC Kinase Signaling Inhibitor 1, SRCIN1, and SNAP25, and further to the SYT-gene family as well as to the DOC2A gene on chromosome 16p11.2-region (Figure 1; www.genecarsd.org). The SRCIN1 protein is part of the cytoskeleton, and the function leads to impaired cell spreading and migration, and it is also involved in exocytosis. It is not normally expressed in fetal bladder. The SNAP25 gene is expressed during the fetal bladder development.
The 16q24.3 deletion (91 kb), contains the gene for Fanconi anemia, complementation group A (FANCA), and the gene Spire-type Actin Nucleation Factor 2, SPIRE2. Fanconi anemia is a clinically heterogenous autosomal recessive disorder associated with congenital malformations in about 70% of the patients. However, no association has previously been described with bladder exstrophy. Our patient was a male with CBE. The deletion was maternally inherited. The SPIRE2 gene is not very well characterized but is involved in intracellular vesicle transport along actin fibers. Similar chromosome aberrations have been reported in other databases.
One male born with CBE carried a paternally inherited deletion on chromosome 18q12.3 (411 kb), involving only the two genes Ras Like Without CAAX 2, RIT2, and Synaptotagmin 4, SYT4. The RIT2 gene is a small GTP-binding protein in the Ras family and is expressed in fetal bladder during the fetal Weeks 5–10 at low levels. Interestingly, the RIT2 gene is also associated with two earlier identified BEEC-genes, the Leucine Zipper Like Transcription Regulator 1, LZTR1 gene on chromosome 22q11.2 and the ISL Lim Homeobox 1, ISL1, gene (GeneCards, Zhang et al., 2013). The ISL1-gene can modify POU4F1 binding sites in the RIT2 promotor region and modulate the activity of the POU4 factors. The POU4F1 is a highly conserved transcription factor expressed in the early embryo.
The SYT4 gene is expressed in neural tissue and in fetal bladder. There is an association with the SYT 1 and 4 with the PPRT2 and DOC2A- genes, located in the 16p11.2 deleted region, through the Synaptosomal-Associated protein 25, SNAP25, gene. The function is membrane fusion and exocytosis. The SNAP25 gene is one of three SNARE proteins in the Golgi apparatus, the others being syntaxin 1A, STX1A, and synaptobrevin or vesicle-associated membrane protein 2 (VAMP2), all three connected to the SYT1 gene. Recently an association to the SYT1 gene, on chromosome 2q21.2, was identified in a Genome Wide Association Study (GWAS) of BEEC (Reutter et al., submitted).
One 50 kb duplication that only covered one single gene, the Chloride Intracellular Channel 6, CLIC6, was detected on chromosome 21q22.12. This gene is expressed in fetal bladder. The protein is located in the plasma membrane and extracellularly. There is an association with the genes YES1 and FYN and thereby also with three POU-genes (POU2F3, POU4F1, and POU5F1), and the SRCIN1 gene, mentioned due to an association with the KIAA1217 gene on chromosome 10.
The Xq23 deletion (50 kb) has not been reported earlier and covers only one single gene, Leucine Rich Repeats and Calponin Homology Domain Containing 2 (LRCH2). The deletion was detected in a male with CBE and is maternally inherited, which may be consistent with an X-linked recessive trait. The gene is highly conserved from Drosophila and regulates cytoskeleton scaffolding (Foussard et al., 2010). The protein interacts with both a family of Latrophilins (LPHN1-3) LPHN1-2-3 and the Dedicator of cytokinesis protein 8, DOCK8, and the Actin-related protein 2/3 complex subunit 3, ARPC2, genes. Both the LPHN2 gene (1p31.1) and the ARPC2 gene are highly expressed in fetal bladder. The LPHN2 gene or Adhesion G protein-coupled receptor L2, ADGRL2 gene is located on chromosome 1p31.1 and its function involves cell adhesion and exocytosis. The network includes Fibronectin Leucine Rich Transmembrane Protein, FLRT1, 2, and 3 and the Leucine Rich repeat Containing G Protein-Coupled Receptor 5 (LGR5), gene that is a link also to WNT-signaling via R-spondins, RSPOND1 and 2.
3.5 Additional findings of unknown significance
Some further findings were evaluated as likely benign given current knowledge based on findings in other disorders or malformations in a clinical setting. They could however still be interesting as parts of different networks or in the pathogenesis of BEEC.
In the deleted region on chromosome 2q35, the Serine/ Threonine protein Kinase 16 gene, STK16, is located. It is expressed in the Golgi apparatus and involves vesicle trafficking. The deleted region on chromosome 3 (3q21.1) contains the SEC22 Homolog A, Vesicle Trafficking Protein gene, SEC22A. This gene is also expressed in the Golgi apparatus and the endoplasmatic reticulum, functions in vesicle trafficking and is associated both with SNAREs complex and other Golgi-located proteins (String network; Szklarczyk et al., 2021). It is further expressed in fetal bladder.
One girl with isolated CBE carried a small 4p16.3 deletion inherited from the father and encompassing only the ZNF141-gene. This region is part of the deleted region in Wolf-Hirschhorn syndrome (WHS; Battaglia et al., 2015). The smallest overlapping region is published as 2.6 Mb, involving almost 50 genes including the ZNF141-gene (Yang et al., 2016). About one third of WHS patients also have urogenital malformations (Battaglia et al., 2008). An association between BEEC and WHS has been described in one case with epispadias and in one among 34 patients with CBE (Nicholls & Duffy, 1998; Schwanitz & Grosse, 1973). The importance of this continuous gene syndrome in bladder disorders was further supported by the finding of somatic FGFR3 mutations in one 8 years old WHS patient with hematuria and a rare form of myofibroblastic bladder tumor (Marte et al., 2013). The region is also important in bladder cancer progression (Bell et al., 1996; Bell et al., 1997; Cappellen et al., 1999; di Martino et al., 2013; Elder et al., 1994; Rothman et al., 2010; Sibley et al., 2000). Mutations in the ZNF141 gene have been described in one family with polydactyly and it is regarded as a tumor suppressor gene (Kalsoom et al., 2013). Comparable small deletions of ZNF141 were detected in 0.02% of 20,000 healthy controls.
The Sidekick cell Adhesion Molecule 1 (SDK1), gene in the duplicated region on chromosome 7p22.2 is expressed in fetal bladder and is associated both with SNAREs complex and carry binding sites for the POU6F1 gene. The main function is reported to be tricellular adhesion (Letizia et al., 2019). Several deletions are reported in public databases Another potentially interesting finding is the duplication on 9q31.1 involving the Syntaxin 17, STX17, gene, active in membrane fusion and associated with SNAREs proteins and Golgi proteins such as Golgi SNAP receptor complex member 1, GOSR1. Similar array findings have been reported twice in public databases.
4 DISCUSSION
We have previously identified altogether five typical 22q11-duplications associated with BEEC (Lundin et al., 2010, 2019). The prevalence in our material is 3.4% in reported BEEC patients as compared with 0.08% in controls (Lundin et al., 2010). We have also reported one family with an X-chromosome rearrangement (Soderhall et al., 2014). Now we have extended the analysis with CMA, using DNA from a further 140 BEEC patients, to try and identify other candidate regions for BEEC. Submicroscopic duplications/deletions possibly associated with BEEC were identified in 16 individuals (11.4%). Earlier studies using CMA have detected novel possibly pathogenic microduplications/ deletions in 9% (10 of 110) and 8% (13 of 169), respectively (Draaken et al., 2013; von Lowtzow et al., 2016). Recently, data on exome sequencing in trios with BEEC were published reporting three gene variants in the WNT3 gene and in the 22q11-duplicated region (LZTR1 and CRKL genes), together with new candidate genes and CNVs (Pitsava et al., 2021).
What is most striking is that studies so far have very few overlapping findings and our data on CNVs show almost no overlap within or compared with other studies. That mutations have been reported in several different genes supports the notion that BEEC has a complex genetic background with the involvement of many different genes as well as possibly environmental factors involved (Reutter et al., 2016). This fits well with the epidemiological data and increased recurrence risks, that do not fit Mendelian inheritance. It is therefore possible that several different gene variants in the same patient may be risk factors. In that case, it would be expected that genetic aberrations detected in affected individuals would be inherited, since one mutation alone may not suffice to cause the disease. Instead, the sum of genetic events inherited from the two parents, or de novo events, sometimes combined with an environmental factor could cause the malformation. It is further likely that genetic variants are instead linked via different molecular pathways that can be disrupted during development, causing BEEC. As outlined in the result section, genes that are identified in this study are associated in networks and pathways. All genes that we highlight here are in addition expressed in human fetal urinary bladder tissue, indicating a role in bladder development and consistent with a role in BEEC pathogenesis.
4.1 Novel findings
Five findings were novel and unique in our study, 1p36.11, 6q23.2, 18q12.3, 21q22.12, and Xq23, and therefore may be of particular interest. The 1p36.11 deletion involves genes that are active in both muscle and skeleton including the ZNF593 gene which negatively regulates POU2F2, part of the Pit-Onc-Unc (POU)-gene family (Hayes et al., 2008; Terunuma et al., 1997). The POU protein family of transcription factors originates from Pit-1, Oct-1, and Oct-2, that are highly conserved from Drosophila, and consists of 6 classes and 15 genes. Their fundamental function is to orchestrate embryonic development and to direct cellular fate decisions (Malik et al., 2018). Thirteen of the POU-genes are expressed in fetal bladder, apart from the POU1F1 and POU4F3, with POU2F1 being the most highly expressed early in development in ectoderm and epidermis. Interestingly, genes in two of the CNVs, 3q26.1, 18q12.3, are associated with the POU networks. Finally, mutations in three different BEEC genes (the ISL1, UPK3A, and LZTR1) have been reported earlier and they are also associated with this network. Such data suggest that especially the POU2F and POU4F networks may be important during fetal bladder development. The TCF21 gene (6q23.2) is active in the embryonal mesoderm surrounding the genitourinary system and is also involved in bladder cancer (Lotfi et al., 2021). Other novel candidate genes are the RIT2 and SYT4 genes and the CLIC6 as well as the LRHC2 gene (Xq23). The latter links to latrophilins (LPHN1-3), a family of cell-adhesion receptors, and WNT signaling pathways. The LPHN1-3 gene family is expressed in fetal bladder, with especially high levels of LPHN2, and these genes are also linked to the gene LLRC37A2 that is listed as a candidate gene to BEEC (MalaCards).
4.2 Findings partly overlapping with earlier studies; the MCC-gene, WNT gene involvement, and the 16p11.2 chromosome region
The MCC gene was involved in two patients, one duplication and one deletion involving different parts of the gene. This gene has previously been reported associated with BEEC due to the location within a larger paternally inherited duplicated region that also included five other genes (von Lowtzow et al., 2016). We have no in-house findings although a few have been reported in public databases. The gene is highly expressed in the human fetal bladder and is active in regulation of the WNT signaling pathway, thus highlighting the importance of WNT in urogenital development. Homozygous WNT3 stop mutations (Q83X) were reported in a large consanginous family with tetra-amelia in three fetuses (Niemann et al., 2004). In two of these, hypoplasia of the pelvis was detected, and one also had persistence of cloaca in addition to other malformations. We have previously reported a de novo WNT3 mutation associated with bladder exstrophy which caused cloaca malformation in zebrafish (Baranowska Körberg et al., 2015). In addition, the first GWAS conducted on BEEC showed an association to a region near the WNT3 and WNT9b genes (Reutter et al., 2014). According to GeneCards, other genes possibly involved in these networks are the CSNK1E, RSPO1-4, and LGR5-genes.
The 16p11.2 deletion syndrome (523 kb) in our study is similar to the duplicated CNV published by Pitsava et al. (2021) which was 733 kb and partly overlapping. At least in individuals carrying 16p11.2 deletions, urogenital malformations are common.
4.3 Associations with genes in the 22q11-region
In the implicated 22q11-region, the genes that most likely function as risk factors for BEEC according to the smallest region of overlap are the CRKL, AIFM3, LZTR1, and THAP7 genes (Draaken et al., 2014). The LZTR1 gene is mainly localized in the Golgi apparatus and helps stabilize the Golgi complex. We have earlier reported a mutation in the LZTR1 gene when an inactivating mutation in LZTR1 gene resulted in defective exocytosis (Lundin et al., 2019). One of the two genes presented here in the 18q12.3 deletion is the RIT2 gene. This gene is novel as a candidate for BEEC. It is expressed in fetal bladder but is also associated with two earlier recognized BEEC genes, both the LZTR1 gene in the 22q11 duplication region, together with the paralog RIT1, as well as the ISL1 gene. The RIT2 gene is neuron specific in mouse retina cells, and it was shown that its promotor was regulated by POU4 TFs, but in addition, ISL1 could modulate this activity (Zhang et al., 2013). The RIT2 gene is also associated with the POU-family genes POU4F1 and POU4F2.
The LRCH2 gene in the Xq23 deletion functions to stabilize cell cortex during cell division as a cytoskeletal scaffolding protein (Foussard et al., 2010). From an earlier WES, we found one frameshift mutation in this gene in one patient with CBE (Baranowska Körberg et al., 2015, unpublished data). The gene is associated with the genes LPHN1-2-3 and in a network with the DOCK8 gene as well as the CRKL gene on chromosome 22.
4.4 Involvement of the Golgi complex
The Golgi apparatus is a central intracellular membrane-bound organelle and involved in both trafficking, cell polarity, and signaling in the cell. Proteins produced in the ER must be compartmentalized and transported in vesicles within the Golgi apparatus, to the cytosol, or to the cell membrane for secretion (Ravichandran et al., 2020; Sun et al., 2020; Tang, 2021). This process is handled by many proteins in complexes called Soluble N-ethylmaleimide-sensitive factor attachment protein receptor, SNAREs, that mainly include the Vesicle Associated Membrane Protein 2 (VAMP2), Synaptosome Associated Protein 25 (SNAP25), and Syntaxin1A (STX1A), but also Syntaxin-5 (STX5), Golgi SNAP Receptor Complex member 1 and 2 (GOSR1, GOSR2), SEC22 Homolog B, Vesicle Trafficking Protein (SEC22B), and Bet1 Golgi Vesicular Membrane Trafficing Protein (BET1). One other important function for the Golgi apparatus is to direct cell polarity while positioning in relation with the nucleus. This function is for example crucial during migration (Ravichandran et al., 2020). As discussed above, many genes are involved in the Golgi function such as the LZTR1, SYT4, SYT1, SEC22A, and KIAA1217, as well as genes in the 16p11.2 deletion (DOC2A, PPRT2).
In addition to the chromosomal findings described in this article we want to emphasize that the task to evaluate pathogenicity of these CNVs is still challenging. CMA is routinely used in clinical genetic diagnostics, which improves the quality of the evaluation of the analysis and findings are evaluated against a growing number of samples. The strength of this study is that it is large, with well-characterized phenotypes. A limitation is that we did not have parental DNA in all patients. Thus, given disparate findings, further studies are needed to understand the pathogenesis of BEEC.
In conclusion, in this study, we performed CMA on DNA from 140 BEEC patients and found possibly pathogenic CNVs in 16 cases. Previous studies and this study have found inactivating mutations or CNVs involving several different genes expressed in human fetal bladder. The number of genes is consistent with a polygenic mechanism for the malformation. We wanted to highlight genes that seem to be involved in networks critical for early human urinary bladder development. Extensive mutation screening has so far only been reported in a limited number of patients, and one would expect that future extensive sequence analysis of BEEC patients and parents will shed more light on the mechanisms behind BEEC. In clinical praxis, it would be advantageous if CMA was performed on all children born with BEEC, both given that around 10%–15% will have a finding that could have clinical relevance and that this will further increase our knowledge about the pathogenesis.
AUTHOR CONTRIBUTIONS
Project management, initiation of the study, and funding: Agneta Nordenskjöld. Planning: Agneta Nordenskjöld and Johanna Lundin. Case recruitment and phenotype characterization: Agneta Nordenskjöld, Samara Arkani, Magdalena Fossum, Magnus Anderberg, Gillian Barker, and Gundela Holmdahl. Experimental work: Johanna Lundin, Jia Cao, Maria Pettersson, and Johanna Winberg. Data analysis: Johanna Lundin, Samara Arkani, and Agneta Nordenskjöld. Writing article draft: Agneta Nordenskjöld, Johanna Lundin, and Samara Arkani. Input on article: all authors.
ACKNOWLEDGMENTS
First, we would like to thank all participating families. We also thank Christina Clementson Kockum who was highly involved in the start of the study and Christina Nyström for technical assistance during the years of collecting DNA samples. The authors would like to acknowledge support from Science for Life Laboratory, the National Genomics Infrastructure, NGI, and Uppmax for providing assistance in massive parallel sequencing and computational infrastructure on fetal expression data.
FUNDING INFORMATION
Swedish Research Council (grant K2012-64X-14506-10-5 2016–01642, to Agneta Nordenskjöld), Foundation Frimurare Barnhuset Stockholm, Stockholm City Council, Karolinska Institutet, the Swedish Brain Foundation, the Harald and Greta Janssons Foundation, and Erik Rönnbergs Foundation.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.




