In This Issue
SALIVA AS ALTERNATIVE TO BLOOD FOR INVESTIGATING AUTISM
Autism spectrum disorder (ASD) is a collection of clinically and genetically heterogeneous disorders thought to be caused by a combination of genetic and environmental factors. Although the genetic etiology remain unclear, a genetic cause can be identified in approximately 30–40% of autistic individuals. Previous large-scale, genome-wide, copy number variation (CNV) studies have consistently shown that CNVs are enriched in autism as compared to control groups. Chromosomal microarray analysis (CMA), generally used for investigating autism, utilizes blood DNA. However, blood collection can be challenging for autistic children, particularly those with repetitive behaviors and sensory and communication issues. In such instances, blood collection may require physical restraint or sedation.
Noninvasive saliva collection offers an alternative to blood collection, but to date, there have been no published studies evaluating saliva DNA and CMA in autism. Wright et al (p. 2913, 10.1002/ajmga.63400) have now evaluated saliva DNA for single-nucleotide polymorphism (SNP) CMA in children with ASD. For the study, saliva DNA was collected from 48 probands and parents (n = 133) with a mean concentration of 141.7 ng/ μL, and SNP CMA was successful in 131/133 (98.5%) of the participants for whom the size and accuracy of a copy number variant(s) called between a proband and carrier parent was correlated, and for a subgroup of 17 probands who had a previous CMA using blood samples.
No discordant copy number variant results were observed between the proband and carrier parent, or the subgroup, although an acceptable mean size difference of 0.009 and 0.07 Mb, respectively, was noted. “Our findings demonstrate that saliva DNA can be an alternative for SNP CMA in autism, which avoids blood collection with significant implications for clinical practice guidelines,” write the authors.
LONG-TERM SEQUENCING MAY HELP IDENTIFY RARE DISEASES
3-Methylglutaconic aciduria is a heterogenous condition that is caused by variants in the CLPB gene (OMIM #616254). It consists of subtypes VIIA and VIIB. Type VIIA is an autosomal dominant condition, while VIIB is an autosomal recessive trait. Characteristics observed in individuals with CLPB deficiency include cataracts, neutropenia, and neurologic involvement with various degrees of severity.
Farrow et al (p. 2908, 10.1002/ajmga.63365) have reported on a family with 2 sons who had biochemical findings of 3-methylglutaconic aciduria and phenotypic features consistent with 3-methylglutaconic aciduria type VIIB. Clinical exome and genome sequencing was completed and nondiagnostic. The family was offered enrollment in a research program that uses HiFi sequencing for unsolved cases of rare disease. HiFi sequencing was completed on both children, and short-read genome sequencing was performed on the parents. HiFi genomes were sequenced to a depth of 30x on a PacBio Sequel IIe. Parental samples were sequenced using TruSeq PCRFree genomes. Parental short-read genome sequencing were sequenced to a depth of 20x on an Illumina NovaSeq 6000.
A previously reported missense variant, CLPB, NM_030813.6: c.491A>C was detected in both children and in the mother's sample, and the intronic variant was predicted to create a novel splice site by multiple algorithms. Full length Isoform Sequencing then confirmed the creation of a cryptic splice site in cmh004696-01 and cmh004696-03, leading to the insertion of 7 amino acids.
“Long-term sequencing, currently performed on a research basis, will likely soon be offered clinically and is expected to play a role in challenging cases where traditional sequencing only identifies one variant,” write the authors.
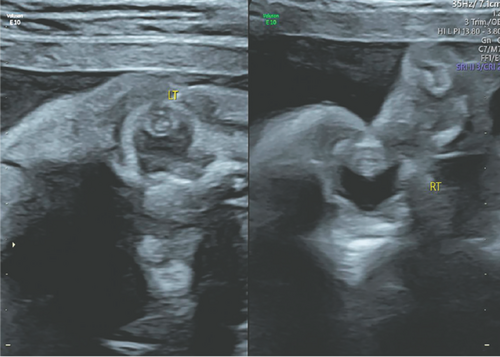
ETHICAL CONSIDERATIONS IN INCIDENTAL FINDINGS FROM PRENATAL EXOME SEQUENCING
Prenatal exome sequencing is used to provide rapid and accurate diagnosis of fetal structural anomalies, but one of its main challenges is how the comprehensive nature of genomic data can yield results that are unrelated to the immediate clinical question. Rudd et al (p. 2856, 10.1002/ajmg.a.63372) report on a case in which the fetus was found to have bilateral congenital cataracts. This condition may be caused by intrauterine infection (CMV, HSV, VZV, toxoplasmosis, rubella, and syphilis), metabolic disorders, and monogenic disorders.
The parents of the fetus decided to undergo whole-exome sequencing, which resulted in an incidental finding of a pathogenic variant in the SCN1A gene unrelated to the bilateral cataracts. The authors note that pathogenic variants in the SCN1A gene are strongly associated with severe myoclonic epilepsy of infancy, or Dravet syndrome, and there is no known association between variants in this gene and congenital cataracts. Based on this finding and after undergoing extensive genetic counseling, the couple decided to terminate the pregnancy at 28 weeks’ gestation.
“This case highlights some of the important clinical and ethical considerations in prenatal genetic diagnosis, particularly in the group of patients in which there is no phenotypic evidence in-utero of the incidental finding,” write the authors. “The case demonstrates the value of frameworks and guidelines to guide management decisions for both clinicians and parents.”