Loeys–Dietz syndrome caused by 1q41 deletion including TGFB2 is associated with a neurodevelopmental phenotype
Abstract
Loeys–Dietz syndrome (LDS) is a connective tissue disorder that commonly results in a dilated aorta, aneurysms, joint laxity, craniosynostosis, and soft skin that bruises easily. Neurodevelopmental abnormalities are uncommon in LDS. Two previous reports present a total of four patients with LDS due to pure 1q41 deletions involving TGFB2 (Gaspar et al., American Journal of Medical Genetics Part A, 2017, 173, 2289–2292; Lindsay et al., Nature Genetics, 2012, 44, 922–927). The current report describes an additional five patients with similar deletions. Seven of the nine patients present with some degree of hypotonia and gross motor delay, and three of the nine present with speech delay and/or intellectual disability (ID). The smallest deletion common to all patients is a 785 kb locus that contains two genes: RRP15 and TGFB2. Previous studies report that TGFB2 knockout mice exhibit severe perinatal anomalies (Sanford et al., Development, 1997, 124, 2659–2670) and TGFB2 is expressed in the embryonic mouse hindbrain floor (Chleilat et al., Frontiers in Cellular Neuroscience, 2019, 13). The deletion of TGFB2 may be associated with a neurodevelopmental phenotype with incomplete penetrance and variable expression.
1 INTRODUCTION
LDS is a connective tissue disorder caused by pathogenic loss-of-function variants in one of six possible genes: TGFBR1, TGFBR2, SMAD3, TGFB2, TGFB3, and SMAD2 (Loeys and Dietz, 2018). Mutations in the transforming growth factor beta 2 (TGFB2, MIM #190220) gene cause LDS type 4 (LDS4; MIM #614816; Lindsay et al., 2012). This gene is located at chromosome band 1q41 and produces a protein that is important for the formation and integrity of the extracellular matrix. TGFB2 regulates cell growth, differentiation, motility, and apoptosis by binding to extracellular receptors, which then activate signaling cascades within the cell.
According to Loeys and Dietz (2018), individuals with LDS typically present with vascular anomalies, such as dilation or dissection of the aorta and other arteries, aneurysms, and vessel tortuosity. Skeletal abnormalities such as pectus excavatum or carinatum, scoliosis, joint laxity, arachnodactyly, osteoarthritis, and osteopenia are also common. Hypertelorism, blue-tinted sclerae, a bifid uvula, cleft palate, and craniosynostosis are also described. Affected individuals typically have soft, translucent skin and may experience easy bruising. Developmental delays, hypotonia, and behavioral issues are considered uncommon findings in patients with LDS.
The prevalence of LDS4 caused by a TGFB2 deletion is unknown, but is suspected to be rare (Loeys and Dietz, 2018). There are two reports describing a total of four individuals with LDS4 caused by pure 1q41 deletions involving TGFB2 (Gaspar et al., 2017; Lindsay et al., 2012). Three of these four previously reported individuals are described with some degree of hypotonia and gross motor delay. In this report, we describe five more individuals with pure 1q41 deletions including TGFB2, four of which present with a neurodevelopmental phenotype of hypotonia, gross motor delay, intellectual disabilities (ID), and/or speech delay.
2 RESEARCH METHODS
2.1 Editorial policies and ethical considerations
This project received expedited approval from Indiana State University's Institutional Review Board (IRB), and IRBs from contributing institutions granted approval for participation when applicable. Individuals with pure 1q41 deletions involving TGFB2 were located through email correspondence with genetic counselors associated with various specialty clinics and hospitals. Patients and/or parents were contacted to provide informed consent for the study. Verbal and written informed consent was obtained for all patients presented in this report.
The data contained in this report were obtained from existing medical records. The genetic providers associated with each patient's care completed a spreadsheet with genotype and phenotype information, such as skeletal, craniofacial, and cardiovascular findings as well as imaging studies and developmental milestones (see Appendix S1). The data contained in the spreadsheets were then used to compare with the other patients as well as the cases previously reported in literature. Chromosomal microarray results interpreted with a prior genome build were updated to GRCh37 nomenclature. For patients #1, #2, and #4, aortic measurements were obtained via two-dimensional (2D) echocardiography. We were unable to ascertain how aortic measurements were obtained on the other patients.
3 CASE REPORTS
Patient #1 is a 12-year-old male with a history of club feet, persistent dislocation of both elbows, recurrent fractures, global developmental delays, autism spectrum disorder (ASD), hypotonia, and spastic cerebral palsy. He was not able to walk until 5 years old. He continues to be ataxic with frequent falls. He vocalizes, but has no specific words. He has feeding difficulties with frequent gagging on textures. He engages in rumination, as well as other self-stimulating and self-injurious behaviors. He experiences sleep disturbance and suspected absence seizures which have not been confirmed with EEG due to poor patient cooperation. His fractures have included both elbows, the right distal humerus, and a metatarsal, all with no known injury. He also has history of a Morgagni congenital diaphragmatic hernia, chronic constipation, multiple dental caries, hyperopia, astigmatism, exotropia, and low thyroid function. On exam he has brachycephaly with a sloped forehead, midface retrusion, ptosis, downslanting palpebral fissures, pectus carinatum, a barrel-shaped chest, and clubbed feet with contractures.
His echocardiogram findings include a dilated Sinus of Valsalva (SV) (Z-score 4.0) and ascending aorta (Z-score 2.9). MRA of the head and neck revealed tortuosity of the internal carotid, cavernous carotid, and basilar arteries. A bone densitometry scan identified mild osteopenia with femoral and femoral neck Z-scores of −1.6 to −2.2, respectively. Chromosomal microarray identified a 7.84 Mb deletion at 1q41 (215,199,578–223,035,427) [GRCh37]. Whole exome sequencing (WES) revealed variants of unknown significance in NRXN1 (p.N365S, c.1094A > G) and PFH2 (p.P1043S, c.3127C > T), but these variants were not believed to be consistent with the patient's phenotype.
Patient #2 is a 22-year-old male with a history of joint hypermobility, recurrent fractures, global developmental delays, and hypotonia. He walked at 18 months old, and he began combining words when he was about 3 or 4 years old. He has attention deficit hyperactivity disorder (ADHD) and anxiety. He experienced sensory integration issues when he was a child. He engages in skin picking behavior and has anger outbursts. He has a seizure disorder and migraines with cyclic vomiting. Fractures have occurred in his leg, clavicle, foot, and wrist. He also has a history of a high and narrow palate, dental crowding, myopia, and a hiatal hernia. On exam, he has dolichocephaly, malar hypoplasia, a tubular nose, a broad and bulbous uvula, pectus carinatum, kyphoscoliosis, bilateral fifth finger clinodactyly, translucent skin, easy cutaneous bruising, and soft, velvety skin.
His echocardiogram findings include a normal SV (Z-score 0.92) and a normal ascending aorta (Z-score 1.23). MRA of the head and neck revealed tortuosity of the carotid, vertebral, and left posterior communicating arteries. A bone densitometry scan revealed diffuse osteopenia. Chromosomal microarray identified a 1.44 Mb deletion at 1q41 (217,589,671–219,026,274) [GRCh37]. His FMR1 repeat analysis for Fragile X was normal. In addition, karyotype identified a 1;6 translocation, but chromosomal microarray did not identify any copy number variants besides the stated 1q41 deletion.
Patient #3 is a 2-year-old female with a history of gross motor delay and hypotonia. She had difficulty learning to crawl and walked at 2 years old. There are currently no concerns about her speech abilities. She has myopia as well as mild turricephaly, downslanting palpebral fissures, ptosis, pectus excavatum, arachnodactyly, and flat feet.
An echocardiogram revealed a PFO, and no further vascular imaging was pursued. Chromosomal microarray identified a 3.99 Mb deletion at 1q41 (216,243,817–220,231,236) [GRCh37]. She also had normal myasthenia panel testing.
Patient #4 is a 43-year-old male with a history of thoracoabdominal aorta dissection and hypermobility. He did not experience any gross/fine motor or speech delays. He has a personal history of a single thumb fracture, hernia, and pneumothorax. He was diagnosed with myopia and later experienced hyperopia. On exam he was found to have dental crowding, scoliosis, pectus excavatum, flat feet, striae, and soft, velvety skin.
His echocardiogram findings include a dilated SV (Z-score 3.4). Further vascular imaging revealed an internal carotid artery dissection, aneurysm of the superior mesenteric artery, ectasia of the left common iliac artery, and tortuosity of the aorta, iliac arteries, and arteries of the neck. Chromosomal microarray identified a 2.17 Mb deletion at 1q41 (217,219,51–219,385,296) [GRCh37]. An aneurysm and dissection panel revealed a variant of unknown significance in COL3A1 (c.3938A > G), which was inherited from a healthy father. WES was also completed but no additional significant variants were found.
Patient #5 is a 7-year-old male with a history of pes planus, a left distal humerus fracture, global developmental delays, ADHD, and ASD. He walked at 2 years old and he spoke his first word at 3 years old. At age 4 years and 6 months, his developmental quotient was calculated as 63. On exam, he has mild generalized joint hypermobility, dolichocephaly, downslanting palpebral fissures, hooded eyelids with supraorbital fullness, micrognathia, and soft, velvety skin.
Echocardiogram at age 5 years identified a borderline dilated SV (Z-score 2.2). An MRA of the abdomen was normal. Chromosomal microarray identified a 785 kb deletion at 1q41 (218,238,773–219,024,035) [GRCh37]. His karyotype is 46,XY, his FMR1 repeat analysis for Fragile X was normal. WES only identified a heterozygous pathogenic PAH variant (p.Arg243Ter).
4 DISCUSSION
Table 1 compares the clinical findings of the five cases of pure 1q41 deletions presented in this report with the four previously reported literature cases. This table demonstrates the variability and overlap in phenotype with pure 1q41 deletions. Seven of the eight patients experience some form of hypotonia and gross motor delay. Patients #1, #2, and #5 have speech delay and ID. Five out of nine patients report some degree of SV dilation, and five of the seven report joint hypermobility. Three of the six patients have osteopenia and recurrent fractures. Pectus deformity is common in the majority of patients (6/7) as are ocular manifestations such as myopia or hyperopia (6/7). Downslanting palpebral fissures are present in five of eight patients.
| Patient #1 | Patient #2 | Patient #3 | Patient #4 | Patient #5 | Gaspar #1 | Gaspar #2 | Lindsay #1 | Lindsay #2 | |
|---|---|---|---|---|---|---|---|---|---|
| Age | 12 yr | 22 yr | 2 yr | 43 yr | 7 yr | 40 yr | 12 yr | 46 yr | 9 yr |
| Sex | M | M | F | M | M | F | M | M | M |
| Break points [GRCh37] | 215,199,578–223,035,427 | 217,589,671–219,026,274 | 216,243,817–220,231,236 | 217,219,510–219,385,296 | 218,238,773–219,024,035 | 215,963,393–220,705,991 | 215,588,712–222,145,072 | 216,672,181–220,202,575 | |
| Development | |||||||||
| Gross motor delay/hypotonia | + | + | + | − | + | NA | + | + | + |
| Speech delay/ID | + | + | − | − | + | − | NA | NA | NA |
| Vascular | |||||||||
| Aortic dimensions (Z-score) | SV = 4.0 Ascending = 2.9 | No dilationa | No dilationa | SV = 3.44 | SV = 2.2 | No dilationa | No dilationa | SV = 2.8 | SV = 3.0 |
| Artery tortuosity | + | + | − | + | NA | NA | NA | NA | NA |
| Skeletal | |||||||||
| Osteopenia | + | + | − | − | − | + | NA | NA | NA |
| Recurrent fractures | + | + | − | Single digit fracture | Single humerus fracture | + | NA | NA | NA |
| Joint hypermobility | + | + | NA | + | + | + | NA | − | − |
| Pectus deformity | + (carinatum) | + (carinatum) | + (excavatum) | + (excavatum) | − | NA | NA | + | + |
| Ocular | |||||||||
| Vision | Hyperopia | Myopia | Myopia | Myopia, Hyperopia | − | NA | NA | Myopia | Hyperopia |
| Cutaneous | |||||||||
| Easy bruising | − | + | − | − | − | + | + | − | − |
| Soft and/or velvety skin | − | + | − | + | + | NA | NA | NA | NA |
| Craniofacial | |||||||||
| Ptosis | + | − | + | NA | − | NA | NA | − | + |
| Downslanting palpebral fissures | + | − | + | NA | + | − | + | − | + |
| Head shape | Brachycephaly | Dolichocephaly | Turricephaly | − | Dolichocephaly | NA | NA | NA | NA |
- Abbreviations: NA, not available; SV, Sinus of Valsalva.
- a Echocardiogram was completed and there was no evidence of aortic dilation.
The smallest 1q41 deletion common among all subjects is 785 kb (218,238,773–219,024,035) [GRCh37] identified in patient #5, indicated by the blue box in Figure 1. There are two genes in this region: RRP15 (MIM #611193) and TGFB2. Of these, TGFB2 is the only gene with a disease phenotype noted in OMIM.
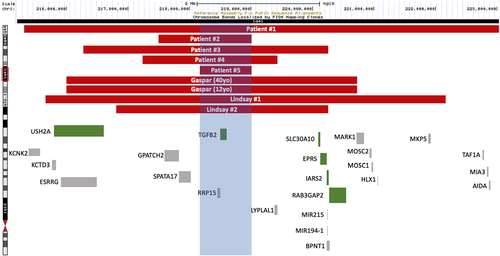
RRP15 is instrumental for pre-60S ribosomal subunit formation, and a reduction in RRP15 protein results in a reduced quantity of 27S and 7S pre-rRNA as well as 5.8S and 25S mature rRNA (de Marchis et al., 2005). The same study found that depletion of RRP15 protein in yeast results in decreased growth. Dong (2017) confirms that the RRP15 protein is involved in ribosome formation, cell growth, and that it plays a role in nucleolus construction as well as cell cycle and checkpoint processes. Current studies are unable to clarify the potential clinical significance of heterozygous RRP15 gene deletions, but it is possible that the loss of this gene may contribute to a neurodevelopmental phenotype through haploinsufficiency.
Sanford (1997) studied TGFB2 knockout mice and reports that the mice exhibit perinatal lethality with several anomalies in multiple organ systems, including lung, heart, skeletal, ocular, inner ear, and urogenital tract. The locations of the anomalies correspond with TGFB2 expression patterns, which led Sanford et al. (1997) to conclude that TGFB2 is involved in epithelial-mesenchymal transformation and that TGFB2 activity is distinctly different from TGFB1 and TGFB3 activity in normal prenatal development.
Chleilat et al. (2019) reports that TGFB2 is active in the embryonic mouse hindbrain floor, which suggests its importance in the formation of hindbrain serotonergic neurons. The same study also suggests that products from other genes, such as TGFB1 and TGFB3, are unable to compensate for deficient TGFB2 product. Furthermore, the authors propose that TGFB2 contributes to neurotransmitter production and uptake as well as involvement at the neuromuscular junction.
Based upon the reports of patients with pure 1q41 deletions involving TGFB2 as well as previous reports on the role of TGFB2 in embryonic development, we propose that the neurodevelopmental phenotype of hypotonia, gross motor delay, speech delay, and ID may be secondary to the deletion of TGFB2. Not all reported individuals exhibit the neurodevelopmental phenotype; even among those who do, the phenotype is varied. This leads us to conclude that a neurodevelopmental phenotype as a result of pure 1q41 deletion involving TGFB2 demonstrates incomplete penetrance and variable expression. It is worth noting that Patient #1 has the largest deletion and is the most severely affected of all reported patients. It is possible that there are other unknown proximal or distal genetic contributions to his phenotype that may be associated with his deletion. It is also striking, however, that patient #2, with a much smaller deletion involving only 5 genes, has a similar severity of developmental delays and behavioral issues.
There are very few reported individuals with pure 1q41 deletions involving TGFB2, therefore resulting in a small sample size. For future studies, more individuals are needed to strengthen the link between the deletion and a neurodevelopmental phenotype. In order to further substantiate the effects of a pure 1q41 deletion involving TGFB2, future research endeavors should investigate other genotype–phenotype associations with LDS4. Contrasting the chromosome deletion with missense or truncation mutations may further characterize the deletion and explain its association with a neurodevelopmental phenotype.
ACKNOWLEDGMENT
We thank the patients and their families for participating in this report.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Deanna Fry interviewed patients and families, compiled data, and drafted the manuscript. Daniel Groepper and Julie Fleischer assisted with data compilation and manuscript drafting. Daniel Groepper, Julie Fleischer, Gretchen MacCarrick, Erin M. Demo, Michael J. Lyons, Margaret J. Wilkes, and Michael J. Lyons contributed patients for inclusion in this study. Megan E. Tucker, Catherine Steding, and Julie Fleischer served on the research advisory committee. All authors edited the manuscript.
Open Research
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in the supplementary material of this article.