Koolen-de Vries syndrome: First report of two unrelated Indian patients
Funding information: Department of Biotechnology, Ministry of Science and Technology, Government of India, Grant/Award Number: BT/PR26428/MED/12/783/2017
Abstract
Koolen-de Vries syndrome is a recurrent microdeletion syndrome caused due to deletion of around 400–600 kb region at 17q21.31. The deleted region contains various genes including KANSL1 and individuals with KANSL1 sequence variations and similar phenotypic manifestations have also been described. It is characterized by typical facial features, variable degree of intellectual disability, seizures, and behavioral problems. Here, we describe the phenotypic features of two patients and to the best of our knowledge, this is the first case report of Koolen-de Vries syndrome from India.
1 INTRODUCTION
Koolen-de Vries syndrome (KdVS; MIM#610443) was first described in 2006. It is characterized by neurological problems such as intellectual disability, speech delay, behavioral issues, hypotonia, seizures, and characteristic facial gestalt including long face, tall broad forehead, upslanting palpebral fissures, epicanthal folds, large ears, and tubular nose with bulbous nasal tip. Structural brain anomalies like ventriculomegaly, corpus callosum aplasia/hypoplasia, hydrocephalus, and joint hypermobility or joint dislocation have also been reported (Sharkey et al., 2009). It is caused due to haploinsufficiency of KAT8 regulatory NSL complex unit 1 (KANSL1) gene either by a microdeletion of chromosomal region 17q21.31 or by sequence variations (Koolen et al., 2012; Zollino et al., 2012). This gene encodes for a nuclear protein that has role in chromatin modification. It is a member of the histone acetyltransferases (HAT) complex (Zollino et al., 2012). The microdeletion ranges from 400 to 600 kb in size and includes C17orf69, CRHR1, SPPL2C, MAPT, STH, and KANSL1 genes. This results from nonallelic homologous recombination between low-copy repeats (Koolen et al., 2008). The prevalence of 17q21.31 deletion is estimated to be around 1 in 55,000 and the exact prevalence of cases due to sequence variation is not known but is expected to be similar to that of microdeletion cases (Koolen et al., 2016). Koolen et al. delineated the phenotypic spectrum of the disease based on the two types of variants reported-microdeletions and KANSL1 variations. The phenotype of the disease is variable in cases with KANSL1 microdeletion as well as those with sequence variants without clear demarcation between the two subsets (Koolen et al., 2016; Zollino et al., 2015).
We hereby report two cases of KdVS syndrome, one with 17q21.31 microdeletion and another with KANSL1 sequence variation.
2 CASE REPORT
2.1 Patient 1
A 5-year-old boy was referred to us with speech delay and hyperactivity. He was the second child of non-consanguineously married healthy parents. He was born at term by caesarean section, due to presence of oligohydramnios. His birth weight was 2.5 kg and there was no history of perinatal asphyxia. At birth, he was diagnosed to have unilateral cleft lip and palate, had associated feeding difficulty, and was operated for the same at 6 months and 1 year of age, respectively. He had delay in attaining gross motor milestones. He started neck holding at 6 months, sitting at 14 months, and walking at 24 months. He has severe speech delay, can speak only bisyllables at present, and indicates his needs by gestures and pointing his finger towards objects. He is toilet trained. His parents have also noticed hyperactivity, overfriendliness with strangers, and poor eye contact. He prefers to play alone and has poor interaction with other children. Also, there is poor response to other sounds and to name-calling. He had recurrent upper respiratory tract infections requiring frequent hospitalizations till the age of 2 years. There is no history of seizures.
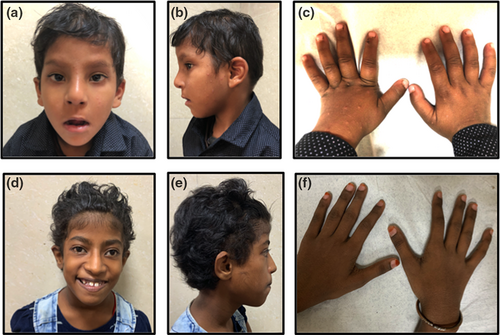
On examination at 5 years of age, his height was 99 cm (−2.37 SD), weight was 15 kg (−1.8 SD), and head circumference was 47.5 cm (−2 SD). Facial features include long face, upslanting palpebral fissures, epicanthic folds, narrow palpebral fissures, broad nasal root, long tubular nose, long philtrum, thin upper lip, and prominent ears with thick ear lobes (Figure 1). Other findings include presence of bilateral broad thumbs. His systemic examination was otherwise unremarkable. His hearing evaluation was normal. Also, no abnormality was detected on abdominal ultrasound, echocardiography, and neuroimaging.
2.2 Patient 2
A 7-year-old girl was referred to us with chief complaint of intellectual disability and bilateral cataract. She was born at term with birth weight of 1.5 kg (at −4.5 SD). Except for low birth weight the perinatal period was uneventful. Hypotonia was documented during her early childhood. Her developmental milestones were delayed. She achieved neck holding at 6 months, sitting at 8 months, and walking at 14 months of age. She started speaking bisyllables at 12 months, some words at 4 years of age, and short unclear sentences after 6 years of age. At the time of her visit, she could make sentences but her speech was not clear. She started imitating others at 6 years of age. She started schooling at 6 years of age and had poor scholastic performance. She likes to play with other children. Also, there is history of recurrent redness in both eyes starting at 5 years for which she was on topical steroids. At 6 years she was diagnosed with bilateral cataract and has been operated for the same. There is no history of seizures or any behavioral abnormality.
On examination (at 10 years of age), her height was 129 cm (−1.5 SD), weight was 26 kg (−1 SD), and head circumference was 51 cm. She had curly hair, low anterior hairline, long face, upslanting eyes, long tubular nose, high arched palate, widely spaced teeth, and prominent ears (Figure 1). She also had dry skin, and joint laxity in her wrist and fingers. She kept her mouth open frequently. No other significant findings were noted in systemic examination. Her echocardiography and neuroimaging were unremarkable.
3 GENETIC ANALYSIS
A written informed consent was taken from patients and family members after initial clinical analysis. Chromosomal analysis at 450-band resolution on peripheral blood leucocytes was performed by cell culture, harvesting, and GTG-banding using standard procedures. DNA was extracted from peripheral blood using QIAamp DNA Mini kit (Qiagen, Hilden, Germany). Multiplex Ligation-dependent Probe Amplification (MLPA) was performed on SALSA MLPA Kit P245 (MRC Holland, Amsterdam, the Netherlands) and analysis was performed using Coffalyser.Net.
Cytogenetic microarray was performed using CytoScan 750K (Affymetrix, Santa Clara, CA) according to the manufacturer's instructions and the result was analyzed using the Chromosome Analysis Suite software (Affymetrix).
Exome sequencing was performed using Illumina sequencing platform following enrichment and capture using Agilent's SureSelect Human V5 (Agilent Technologies, Santa Clara, CA). The sequences reads were aligned to GRCh37/hg19 human reference genome. Bioinformatics analysis was performed using an in-house pipeline based on GATK and annotated using Annovar. Variant prioritization was performed based on population based allele frequency, variant location and function, possible modes of inheritance, and phenotype relevance.
For verification and family studies, primers were designed and PCR was performed followed by Sanger sequencing.
4 RESULTS
4.1 Patient 1
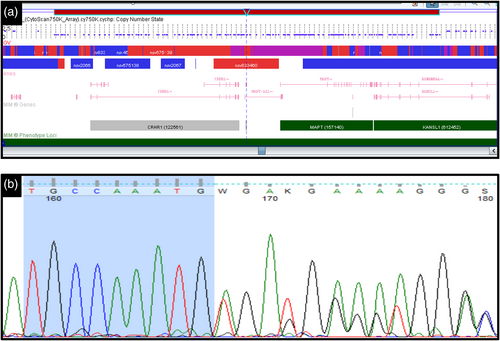
Peripheral blood karyotype was found to be normal (46,XY). MLPA showed heterozygous deletion at 17q21.31. For confirmation, cytogenetic microarray was done and a heterozygous deletion of 556 Kb was detected on 17q21.31 {arr[hg19]17q21.31(43,645,879-44,201,470)x1} encompassing CRHR1, SPPL2C, MAPT, STH, and KANSL1 genes (Figure 2a). This deletion was interpreted as pathogenic according to American College of Medical Genetics and Genomics (ACMG) guidelines. Testing of parents could not be performed.
4.2 Patient 2
Her peripheral blood karyotype was found to be normal (46,XX) and there was no clinically relevant copy number variation on cytogenetic microarray. Exome sequencing identified a 5 bp heterozygous deletion c.2772_2776del; p.Cys924*, in KANSL1 gene (NM_015443). This variant has not been reported in population databases (gnomAD, 1,000 Genomes Project, ExAC browser and in-house data of 720 individuals). It is also not reported in Clinvar, HGMD, LOVD, and Pubmed. The in-silico prediction tools have predicted this variant to be pathogenic. The heterozygous 5 bp deletion was confirmed in the patient on Sanger sequencing (Figure 2b) and was absent in the parents. So, this variant was classified as pathogenic as per ACMG/AMP (Association for Molecular Pathology) guidelines.
5 DISCUSSION
Here, we report two cases of Koolen-de Vries syndrome from India with characteristic features such as facial dysmorphism and psychomotor delay mainly speech impairment. The affected individuals had varying degree of intellectual disability, behavioral issues such as hyperactivity, overfriendliness and attention deficit; language impairment was more as compared to delay in motor milestones. Both of the cases described in this report had developmental delay along with speech delay and patient 1 also had hyperactivity with overfriendly behavior with strangers. Both of them had characteristic facial features described in KdVS including curly hair, long face, epicanthal folds, upslanting palpebral fissures, tubular nose, long columella, high arched palate, and prominent ears. Patient 1 also had cleft lip and cleft palate. In a series by Koolen et al., cleft palate was found in 9% of cases (Koolen et al., 2008). Skeletal abnormalities have been described in around 36% of cases and include scoliosis, kyphosis, hip dislocation, positional deformities of feet. Patient 1 was found to have broad thumbs which have been reported in cases of KdVS syndrome (Barone et al., 2015). Patient 2 had intrauterine growth retardation which has been reported in 26% of cases previously (Koolen et al., 2016). There is no history of seizures in our patients, but in various case series, around 40–50% of patients were having various types of seizure disorders (Dubourg et al., 2011). KdVS may also be associated with some other structural malformations such as central nervous system, ophthalmological, cardiac defects (~40%), and urogenital malformations. Patient 2 had bilateral cataract which has been described previously in cases with KdVS syndrome or in this case may be associated with the prolonged use of topical steroids (Koolen et al., 2008). The central nervous system abnormalities include ventriculomegaly, anomalies of corpus callosum, white matter abnormalities, etc. (Egloff et al., 2014). None of these structural malformations were found in our patients.
At his first visit, patient 1 was suspected to have 22q11.21 microdeletion on the basis of developmental delay, recurrent infections, facial features, that is, long tubular nose with prominent root, cleft lip, and palate, but diagnostic testing by MLPA revealed 17q21.31 deletion which was later also confirmed by array CGH. Also, in a cohort of 41 patients with clinical diagnosis of Di-George syndrome, 12% were found to have other submicroscopic chromosomal imbalances including one case of KdVS (Koczkowska et al., 2017).
The phenotype is highly variable and a study by Koolen et al. showed that there is no significant difference between cases with deletion and those with sequence variation (Koolen et al., 2016). Our patients also had the typical features associated with KdVS.
The novel sequence variation detected in patient 2 further emphasizes the importance of molecular testing such as next generation sequencing in confirmation of diagnosis in cases with intellectual disability. Widespread testing will help in estimation of actual prevalence of this condition and counseling of family.
6 CONCLUSION
This is the first report from India describing two patients with KdVS syndrome. Although the clinical phenotype is highly variable, the dysmorphic features of this syndrome are consistent across various ethnicities. The identification of a new variant in addition to new clinical findings in our patients will aid in increasing our understanding of the clinical and genetic spectrum of the disease.
ACKNOWLEDGMENTS
We thank the patient and the family for their participation in the study. This study was funded by Department of Biotechnology, Ministry of Science and Technology, Government of India (Grant No. BT/PR26428/MED/12/783/2017).
CONFLICT OF INTEREST
None.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




