A rare cause of syndromic short stature: 3M syndrome in three families
Abstract
3M syndrome is a rare autosomal recessive genetic disorder characterized by severe growth retardation, dysmorphic facial features, skeletal dysplasia, and normal intelligence. Variants in CUL7, OBSL1, and CCDC8 genes have been reported to be responsible for this syndrome. In this study, the clinical and molecular findings of four 3M syndrome cases from three families are presented. All cases had growth retardation, relative macrocephaly, and typical dysmorphic facial features. Their neurological developments were normal. Sequencing of CUL7, OBSL1, and CCDC8 genes revealed two different novel homozygous variants in CUL7 in Families 1 and 3 and a previously reported homozygous pathogenic variant in OBSL1 in Family 2. In conclusion, a comprehensive dysmorphological evaluation should be obtained in individuals presenting with short stature and in such individuals with typical facial and skeletal findings, 3M syndrome should be considered. Our report expands the genotype of 3M syndrome and emphasizes the importance of thorough physical and dysmorphological examination.
1 INTRODUCTION
3M syndrome is a rare autosomal recessive disorder, characterized by severe pre- and postnatal growth retardation, dysmorphic facial features, skeletal dysplasia, and normal intelligence (Huber, Munnich, & Cormier-Daire, 2011). Additionally, short neck, square shoulders, sternal abnormalities, narrow thorax, wing scapula, lordosis, joint laxity, short fifth finger, and rocker bottom feet can be associated with this syndrome.
3M syndrome was first described by Miller et al. (Miller, McKusick, Malvaux, Temtamy, & Salinas, 1975) and three genes (Cullin 7, CUL7; Obscurin-like 1, OBSL1; Coiled-coil domain-containing protein 8, CCDC8) responsible for this disorder have been identified (Hanson et al., 2009; Hanson et al., 2011; Huber et al., 2005). CUL7 encodes a large protein which is a member of the cullin family. CUL7 is a structural component of a SKP1-CUL7-FBXW8-ROC1 (SCF) E3 ubiquitin ligase complex that is responsible for ubiquitin-mediated proteasomal degradation (Dias, Dolios, Wang, & Pan, 2002). Recently, Yan et al. (Yan et al., 2014) showed that CUL7, OBSL1, and CCDC8 physically interact with each other and form a complex (the 3M complex) which maintains microtubule and genome integrity. Disruption of the 3M complex via deletion or knockdown of any 3M gene results in severe microtubule damage, abnormal chromosome segregation, and cell death. However, the cellular mechanism underlying 3M syndrome still remains unclear.
Here, we evaluate clinical and molecular manifestations of four individuals with 3M syndrome from three different families, and report two novel variants in CUL7 expanding the molecular spectrum of the syndrome.
2 MATERIALS AND METHODS
2.1 Study group
Four Turkish cases from three different families with short stature and dysmorphological findings consistent with 3M syndrome were included in the study. All cases were examined by an expert clinical geneticist. Demographic data, family history, clinical features, and radiographic findings were all obtained from hospital records.
2.2 Molecular analysis
Sequence analysis of the CUL7 (NM_001168370), OBSL1 (NM_015311), and CCDC8 (NM_032040) genes was performed using Sanger sequencing in Case 1, Case 2, and Case 3. However, in Case 4, a targeted next generation sequencing (NGS) panel (TruSight One Sequencing Panel by Illumina) was used to perform molecular analysis. Nextera XT DNA Library Preparation Kit (Illumina Inc., San Diego, CA) was used for target enrichment; in accordance with the manufacturer's instructions. Paired end sequencing was performed on all samples using the Illumina NextSeq platform (Illumina Inc., San Diego, CA). NGS data was analyzed using Illumina VariantStudio software and IGV (Integrative Genomics Viewer).
The frequencies of the identified variants were investigated in different databases: NCBI dbSNP build141 (http://www.ncbi.nlm.nih. gov/SNP/), 1000 Genomes Project (http://www.1000genomes.org/), Exome Aggregation Consortium (ExAC) (http://exac.broadinstitute.org/), and Genome Aggregation Database (gnomAD) (http://gnomad.broadinstitute.org/). The impact of variants on the protein structure was evaluated using several in silico prediction tools such as SIFT, MutationTaster, and REVEL (Ioannidis et al., 2016; Kumar, Henikoff, & Ng, 2009; Schwarz, Rodelsperger, Schuelke, & Seelow, 2010). Conservation of residues across species was evaluated by GERP (Davydov et al., 2010). American College of Medical Genetics (ACMG) guidelines have been used for the classification of the pathogenicity (Richards et al., 2015). Segregation analysis was performed using Sanger Sequencing.
Samples from the cases were obtained in accordance with the Helsinki Declaration. Written informed consent for genetic testing was obtained from all cases and/or their parents/guardians.
3 RESULTS
3.1 Clinical characteristics of cases
3.1.1 Case 1
Case 1 was referred to our department at 14 months of age due to short stature and his dysmorphic features. He was born to consanguineous parents at 39 weeks of gestation. His birth weight, height and head circumference were 2,500 g (−1.68 standard deviation score (SDS)), 45 cm (−1.85 SDS), and 35 cm (−0.42 SDS), respectively (Gokcay, Furman, & Neyzi, 2008).
On physical examination, his weight, height, and head circumference were 4,410 g (−4.28 SDS), 61.5 cm (−4.82 SDS), and 43 cm (−1.03 SDS), respectively. He was noted to have a triangular face, frontal bossing, depressed nasal bridge, fleshy, upturned nose, anteverted nares and a pointed, prominent chin.
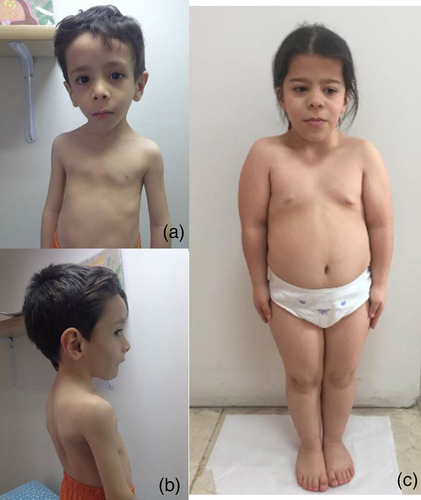
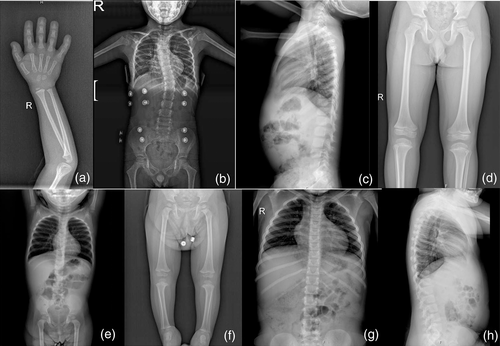
He was reevaluated at 3 years and 5 months of age. His height and annual growth rate were 75.7 cm (−5.85 SDS) and 4.1 cm/year (−2.2 SDS), respectively. He was noted to have pectus carinatum, narrow thorax and hyperlordosis in addition to his previous dysmorphological findings (Figure 1). Testicles, bilaterally, were found to be in scrotal sac and of a prepubertal size. A skeletal bone survey showed delayed bone age, thoracic scoliosis, slender long tubular bones, and mild metaphyseal widening (Figure 2).
Serum basal growth hormone (GH), insulin-like growth factor-1 (IGF1), and IGF binding protein-3 (IGFBP3) levels were 10.2 ng/mL, 52.6 ng/mL (−1.27 SDS), and 2,921 μg/mL (2.78 SDS), respectively (Table 1). Due to normal serum basal GH levels (>7 ng/ml) and peak GH responses in GH stimulation test, an IGF1 generation test (50 μg/kg/day/5 days) was performed. Following stimulation, a sufficient increase in IGF1 level was detected.
| Case 1 | Case 2 | Case 3 | Case 4 | |
|---|---|---|---|---|
| Basal IGF-1 (ng/mL)/SDS | 52.6/−1.27 | 75/−1.12 | 70/−0.44 | 158/1.13 |
| Basal IGFBP-3 (μg/mL)/SDS | 2921/2.78 | 3968/0.69 | 2290/1.11 | 4760/0.59 |
| Basal GH (ng/mL) | 10.2 | 0.30 | 2.47 | 0.95 |
| GH at 30th min. | >40 | 0.27 | 5.33 | 7.73 |
| GH at 60th min. | >40 | 0.167 | 2.04 | 2.00 |
| GH at 90th min. | – | – | 5.39 | 2.64 |
| Baseline height SDS | −5.85 | −3.87 | −5.33 | −5.04 |
| Baseline annual growth velocity (cm/year)/SDS | 4.10/−2.2 | 4.78/−1.52 | 3.2/−3.56 | 1.63/−3.51 |
| GH dosage (mg/kg/day) | 0.035–0.050 | 0.045 | 0.045 | – |
| Height SDS (following GH therapy) | −5.33 | −2.76 | −3.94 | – |
| Annual growth rate (following GH therapy) (cm/year)/SDS | 3.99/−1.9 | 5.95/0.65 | 6.04/−0.56 | – |
- Abbreviation: SDS, standard deviation score.
Although basal GH level was found to be normal, recombinant human GH (rhGH) therapy was given because he was born small for gestational age (SGA), had severe short stature and was failed to achieve catch-up growth. Although a high dose (35–50 μg/kg/day) of rhGH therapy was initiated, treatment was discontinued following 4 years due to low growth velocity (3.5–4.5 cm/year).
He was 12 years old at last examination and his weight was 17.05 kg (−4.74 SDS) and height 109 cm (−5.73 SDS). His intelligence and neuromotor development were normal. He was wearing a corset for his scoliosis.
3.1.2 Cases 2 and 3
Case 2 was the first child of a healthy consanguineous couple. He was born at 39 weeks of gestation, and birth weight was 2,350 g (−3.00 SDS), and height was 43 cm (3.44 SDS). He was considered to have achondroplasia and tested for FGFR3 at the age of 3 months. FGFR3 sequencing revealed no variants.
On physical examination, at the age of 3 and a half years, his weight, height, and head circumference were 12.7 cm (−1.85 SDS), 84 cm (−4.07 SDS), and 52 cm (0.77 SDS), respectively. Rhizomelic short stature, relative macrocephaly, triangular face, frontal bossing, depressed nasal bridge, fleshy and upturned nose, pointed and prominent chin, and oligodontia were observed. He had also hyperlordosis, broad and fleshy hands and feet, prominent heels, and joint laxity. External genitalia examination was found to be normal. His neuromotor development was normal. Skeletal survey revealed tall vertebral bodies and long, slender tubular bones (Figure 2).
At the age of 6 years and 5 months, his height and annual growth rate were 99.7 cm (−3.87 SDS) and 4.78 cm/year (−1.52 SDS), respectively. Serum basal GH levels and peak GH responses to GH stimulation test were found to be low. An rhGH therapy (45 μg/kg/day) was administered due to low GH levels and decreased growth velocity. Treatment is still ongoing, and his recent annual growth velocity was 5.95 cm/year (0.65 SDS). Following treatment period, his height SDS was decreased to −2.76.
Case 3 was the affected sibling of Case 2. She was born at 38 weeks of gestation via caesarean section. Birth weight and length were 2,300 g (−2.12 SDS) and 42 cm (−3.17 SDS), respectively.
On physical examination at the age of 22 months, her weight, height, and head circumference were 7.7 kg (−3.14 SDS), 63 cm (−6.46 SDS), and 46 cm (−1.25 SDS), respectively. Relative macrocephaly, triangular face, frontal bossing, midface hypoplasia, bulbous nose, anteverted nares, pointed, and prominent chin were observed. She had also hyperlordosis, pes planus, prominent heels, and joint laxity. Her neuromotor development was normal. Her skeletal survey was found to be normal.
Serum basal GH level and peak GH responses to GH stimulation test were found to be low. An rhGH therapy (45 μg/kg/day) was administered due to low GH levels and decreased growth velocity (3.2 cm/year, −3.56 SDS) (Table 1). At the age of 5 years, GH therapy is still continuing, and her recent annual growth velocity was 7.6 cm. The baseline height SDS was −6.46, following treatment period, has decreased to −3.94.
3.1.3 Case 4
A 9-year-old girl with short stature was referred to our clinic for differential diagnosis of skeletal dysplasia. She was born at 39 weeks of gestation. Her parents were not consanguineous, however, were from the same small village.
Her weight, height, and head circumference were 29.4 kg (0.06 SDS), 105 cm (−4.69 SDS), 53 cm (0.46 SDS), respectively. She had short stature, relative macrocephaly, depressed nasal bridge, broad nasal tip, and thick lower vermilion. Short neck, wide and flat thorax, square shoulders, and pes planus were observed (Figure 1). A bone survey revealed long and slender tubular bones, schmorl nodules and posterior fusion in L4-5 (Figure 2).
GH stimulation test was performed. Basal GH level was found to be low and a partial GH peak was obtained (Table 1). An rhGH therapy has been planned to be initiated.
In all four cases, complete blood count and biochemical tests for liver, kidney and thyroid functions were found to be in the normal ranges. Tissue transglutaminase IgA was negative. Left wrist X-rays showed delayed bone age by Greulich-Pyle atlas in all cases (WW & SI, 1959). Hypophysis MRI, abdominal ultrasonography, and echocardiography all were normal. Clinical characteristics of cases are given in Table 2. Given the clinical, dysmorphological and radiologic features, 3M syndrome was considered in the cases.
| Family no | 1 | 2 | 3 | |
|---|---|---|---|---|
| Case no | 1 | 2 | 3 | 4 |
| Age (during last examination) | 12 years | 10 years | 5 years | 10 years |
| Age (during the first examination) | 14 months | 3 and a half years | 22 months | 9 years |
| Gender | Male | Male | Female | Female |
| Prenatal findings | IUGR | Short extremities, IUGR | Short extremities, IUGR | Short extremities |
| Birth weight (gr/SDS) | 2,500/−2.49 | 2,350/−3.00 | 2,300/−2.12 | 2,500/−1.99 |
| Birth height (cm/SDS) | ? | 43/−3.44 | 42/−3.17 | 38/−5.52 |
| Height (during the first examination), cm/SDS | 61.5/−4.82 | 84/−4.07 | 63/−6.46 | 105/−4.69 |
| BMI (during the first examination)/SDS | 14.16/−2.32 | 18/1.77 | 19.4/1.75 | 26.67/2.58 |
| OFC (during the first examination), cm/SDS | 43/−1.03 | 52/0,77 | 46/−1.25 | 53/0.46 |
| Facial features | ||||
| Relative macrocephaly | + | + | + | + |
| Dolichocephaly | + | + | + | + |
| Triangular face | + | + | + | + |
| Hypoplastic midface | + | + | + | + |
| Fleshy nasal tip | + | + | + | + |
| Long philtrum | + | + | + | + |
| Full lips | + | + | + | + |
| Pointed chin | + | + | + | + |
| Musculoskeletal features | ||||
| Short broad neck | + | + | + | + |
| Square shoulders | + | + | + | + |
| Short thorax | + | + | + | + |
| Sternal anomaly | + | − | − | + |
| Hyperlordosis | + | + | + | |
| Generalized or isolated joint hypermobility | − | + | + | |
| Prominent heels, and pes planus | + | + | + | + |
| Genitourinary anomalies | ||||
| Hypospadias | − | − | ||
| Hypogonadism | − | − | ||
| Intelligence | Normal | Normal | Normal | Normal |
| Radiographic features | ||||
| Long slender tubular bones | + | + | − | + |
| Tall vertebra | + | + | − | + |
| Wedging of the thoracic vertebral bodies | − | − | − | + |
| Thoracic kyphoscoliosis | + | − | − | − |
| Small pelvis | − | − | − | − |
| Molecular findings | ||||
| Karyotype | 46,XY | 46,XY | 46,XX | 46,XX |
| Gene | CUL7 (NM_001168370) |
OBSL1 (NM_015311) |
OBSL1 (NM_015311) |
CUL7 (NM_001168370) |
| Variation (DNA) | c.571_572delCA | c.1273dupA | c.1273dupA | c.4666dupG |
| Variation (protein) | p.His191Ter | p.Thr425AsnfsTer40 | p.Thr425AsnfsTer40 | p.Val1556GlyfsTer30 |
| Novelty | Novel | Known | Known | Novel |
| ACMG classification | Pathogenic | Pathogenic | Pathogenic | Pathogenic |
- Abbreviations: BMI, body mass index; IUGR, intrauterine growth retardation; OFC, occipito frontal circumference; SDS, standard deviation score.
3.1.4 Molecular findings
In Case 1, molecular analysis revealed homozygous c.571_572delCA variant in CUL7 (NM_001168370) (Figure 3). This small deletion results in a premature termination codon at 191th position of CUL7 mRNA. The variant has not previously been reported in public databases. The GERRP score was 5.5999. Via segregation analysis, both parents were found to be heterozygous carriers
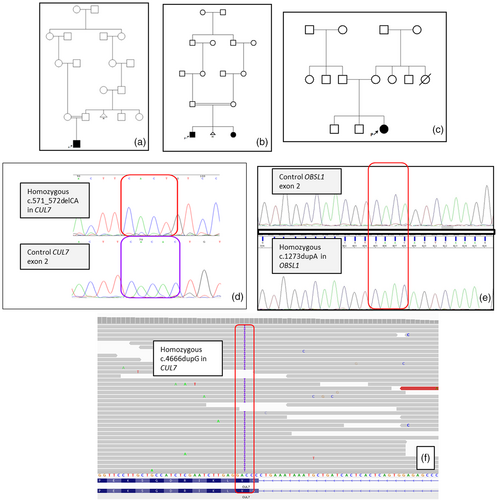
In Cases 2 and 3, a homozygous c.1273dupA variant resulting in a premature termination codon (p.Thr425AsnfsTer40) was detected in OBSL1 (NM_015311) (Figure 3). This variant (rs762334954) with a minor allele frequency of 1/5231 has been reported in gnomAD database, and classified as pathogenic in ClinVar. A number of 3M patients carrying this variant homozygously or compound heterozygously have been reported in the literature (Hanson et al., 2009; Huber et al., 2010; Keskin et al., 2017). Both parents were found to be heterozygous carriers for the same variant.
In Case 4, homozygous c.4666dupG variant in CUL7 was detected (Figure 3). This duplication results in a premature termination codon at 1556th position of CUL7 mRNA. The variant has not previously been reported in public databases. The GERRP score was 5.1617. The parents were found to be heterozygous for the variant.
4 DISCUSSION
The clinical findings and molecular analysis in our four cases confirmed the diagnosis of 3M syndrome. The 3M syndrome is a rare autosomal recessive genetic disorder, with approximately 200 individuals have been reported in the literature thus far (Clayton et al., 2012). It is a phenotypically homogeneous disease, characterized by severe short stature and dysmorphic features. Pathogenic variants can be detected in CUL7 and Obscurin-like 1 (OBSL1) gene in approximately 67–77.5% and 16.3–28% of cases, respectively (Clayton et al., 2012; Huber et al., 2011). The Coiled-coil domain-containing protein 8 (CCDC8) gene variants have been reported rarely (5). In a study by Huber et al., no genotype–phenotype correlation could be established in the syndrome (Huber et al., 2011). However, Simsek-Kiper et al. investigated genotype–phenotype correlation in 24 3M syndrome individuals from 19 unrelated families. They identified disease-causing variants in CUL7 or OBSL1 genes in 75% of the study group (Simsek-Kiper et al., 2019). They reported that cases with CUL7 variants had lower mean birth weight, birth weight SDS, and height SDS at admission than cases with OBSL1 variant. In our study, we have not observed any significant differences of these parameters among the cases with pathogenic variants in CUL7 or OBSL1.
In this study, we identified two novel variants, c.571_572delCA and c.4666dupG, in CUL7 gene in Families 1 and 3, respectively. Both variants were null variants and classified as pathogenic in accordance with ACMG recommendations. Additionally, the variants segregation is compatible with the mode of inheritance of the disease in each family. Therefore, both variants in CUL7 gene were considered to be disease-causing.
In the differential diagnosis of 3M syndrome, Silver Russell syndrome, Floating Harbor syndrome, Bloom syndrome, Mulibrey Nanism syndrome, Seckel syndrome, Dubowitz syndrome and Microcephalic Osteodysplastic Primordial Dwarfism Type 2 should be considered (Clayton et al., 2012; Huber et al., 2011). Individuals with 3M syndrome have normal intelligence as a distinguishing finding from these syndromes. Skeletal defects are also common in individuals with 3M syndrome. If there are specific X-ray findings such as delayed bone age, long and slender tubular bones, flared metaphyses, tall vertebral bodies, thoracic kyphoscoliosis, and spina bifida occulta in a child with short stature, 3M syndrome should be considered in the differential diagnosis (Clayton et al., 2012). A comprehensive dysmorphological evaluation with normal cognitive functions, radiologic manifestations and molecular findings provided a diagnosis of 3M syndrome in our cases.
There is no specific therapy for the disease; however, several studies evaluated the responses to GH treatment in 3M syndrome individuals. It has been reported that GH levels are usually normal in the 3M syndrome individuals (Clayton et al., 2012; Guven & Cebeci, 2011; Huber et al., 2011). Simsek-Kiper et al. found sufficient peak GH responses in 87.5% (seven cases) of eight cases in whom the GH axis was assessed (Simsek-Kiper et al., 2019). Clayton et al. investigated the response to recombinant human GH (rhGH) treatment in 16 individuals (Clayton et al., 2012). They reported a small but significant increase in height velocity SDS and in height SDS following 1 year of rhGH therapy. However, some individuals fail to respond to the GH treatment to stimulate catch-up growth (Demir, Altincik, & Bober, 2013; Guven & Cebeci, 2011). In our study, GH levels were found to be low in Cases 2 and 3. Three cases had received GH. In Case 1, despite normal GH levels, GH treatment was performed. However, following therapy no increase in height velocity was noted. In contrast, a slight increase in height SDS was detected in Cases 2 and 3. It has been considered that severe short stature in 3M syndrome is a consequence of GH resistance (Clayton et al., 2012). Therefore, despite the presence of normal GH levels, a GH therapy is recommended. Duration of the treatment should be decided in accordance with the increase in height SDS and growth velocity.
Because of normal intelligence, prenatal diagnosis of 3M syndrome is controversial. Pre-implantation genetic diagnosis is a more acceptable choice ethically than prenatal diagnosis for families who want to have an unaffected child.
In conclusion, 3M syndrome should be considered in the differential diagnosis of short stature especially when accompanied with dysmorphic features and normal intelligence. Molecular analysis of CUL7, OBSL1, and CCDC8 genes is needed to confirm the clinical diagnosis in suspected cases and to provide appropriate genetic counseling. We report two novel variants in the CUL7 gene, thus expanding the molecular spectrum of 3M syndrome.
ACKNOWLEDGMENT
The authors thank to Daniel Hanson for preforming molecular analysis.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
Research data are not shared.




