De novo variant in AMOTL1 in infant with cleft lip and palate, imperforate anus and dysmorphic features
Abstract
AMOTL1 belongs to the Motin family of proteins that are involved in organogenesis and tumorigenesis through regulation of cellular migration, tube formation, and angiogenesis. While involvement of all AMOTs in development or suppression of cancers is relatively well described, little is known about the congenital phenotype of pathogenic variants in these genes in humans. Recently, a heterozygous variant in AMOTL1 was published in association with orofacial clefts and cardiac abnormalities in an affected father and his daughter. However, studies in mice did not recapitulate the human phenotype and the case was summarized as inconclusive. We present a female infant with cleft lip and palate, imperforate anus and dysmorphic features, in whom trio exome sequencing revealed a de novo variant in AMOTL1 affecting a highly conserved amino acid (c.479C>T; p.[Pro160Leu]). Bioinformatic predictions and in silico modeling supported pathogenicity. This case reinforces the conjecture regarding the disruptive effect of pathogenic variants in AMOTL1 on organ formation in humans. Studies of additional families will reveal the full phenotypic spectrum associated with this multiple malformation syndrome.
1 INTRODUCTION
Angiomotins or Motins (AMOTs) comprise a family of three adaptor proteins that are involved in various mechanisms of cellular migration, tube formation, angiogenesis and both tumor evolvement and suppression (Huang et al., 2018). Angiomotin (AMOT), the first characterized member of the family, was reported in 2001 by Troyanovski et al. who suggested that its mechanism of action is via binding angiostatin, a circulating inhibitor of endothelial cell migration, tube formation and an inducer of cell type-specific apoptosis (Troyanovsky, Levchenko, Månsson, Matvijenko, & Holmgren, 2001). In 2002, Nishimura et al. cloned and characterized the junction-enriched and -associated protein (Jeap) which was subsequently renamed angiomotin like-1 (AMOTL1). AMOTL1 was shown to co-localize with F-actin at tight junctions. It also controls cell polarity and paracellular permeability (Gagné et al., 2009; Nishimura et al., 2002; Zheng et al., 2009). AMOTL2, the third member of the group, was shown to be involved in regulation of the cytoskeleton and tight junctions (Lv et al., 2017; Mojallal et al., 2014). AMOTs share structural homology of conserved glutamine-rich N-terminal domains and coiled-coil and PDZ-binding domains which compose the C-terminal region (Bratt et al., 2002; Moleirinho, Guerrant, & Kissil, 2014). Unlike AMOT, neither AMOTL1 nor AMOTL2 possess an angiostatin-binding domain (Huang et al., 2018). Although AMOTs differ significantly in their spatiotemporal and tissue expression levels (Moleirinho et al., 2014), they appear to have a certain degree of functional overlap. For example, Li et al. (2012) observed that in-vitro overexpression of any single AMOT family member in mammalian cells led to repression of WNT/β-catenin signaling whereas knockdown did not result in overexpression. Only co-knockdown of all three AMOTs resulted in significant overexpression of WNT/β-catenin signaling, suggesting functional redundancy. In addition, AMOTs appear to act as scaffold proteins that promote the Hippo signaling pathway which controls cell proliferation, differentiation and survival by regulating Yes-associated proteins (YAP) (Mana-Capelli & McCollum, 2018; Moleirinho et al., 2014). Overall, this pathway plays an important role in organogenesis and tumorigenesis (Ishihara & Nishina, 2018).
The human AMOTL1 is a 956 amino acid protein with a molecular weight of 106 kilodaltons (Nishimura et al., 2002). The protein is encoded by AMOTL1 located on chromosome 11q21. Human AMOTL1 mRNA is most widely expressed in skeletal muscle tissue and least expressed in the blood (Moleirinho et al., 2014). While involvement of all AMOTs in development or suppression of cancers is relatively well described (Huang et al., 2018), little is known about the congenital phenotypic effect of pathogenic variants in these genes in humans. In 2019, Liegel et al. reported a nonconsanguineous family where the father harbored a de novo heterozygous missense variant in a highly conserved region of AMOTL1 which was inherited by his daughter. Both the father and his daughter had a phenotype of congenital cleft lip and palate and abnormal ear shape. The daughter also suffered from tetralogy of Fallot (TOF) cardiac anomaly, finger contractures and advanced bone age. We present a second case of a predicted pathogenic variant in AMOTL1 and provide phenotypic expansion of the associated clinical spectrum.
2 MATERIALS AND METHODS
2.1 Exome analysis
Following informed consent, exonic sequences from DNA of the proband and both parents were enriched with the SureSelect Human All Exon 50 Mb V5 Kit (Agilent Technologies, Santa Clara, CA). Sequences were generated on a NovaSeq6000 sequencing system (Illumina, San Diego, CA) as 150-bp paired-end runs. Read alignment and variant calling were performed with DNAnexus (Palo Alto, CA) using default parameters with the human genome assembly hg19 (GRCh37) as reference. Exome analysis of the proband yielded a mean coverage of 51X, with more than 90% reads over 20X.
2.2 Sanger sequencing
An amplicon containing the variant of interest was amplified by conventional PCR of genomic DNA using the following primers: AMOTL1_F: 5′-GACCAGCCCATCCTACAAAC-3′ and AMOTL1_R: 5′-AGAACTGCGACTGTGCTTTG-3′. The resultant fragment was analyzed by Sanger dideoxy nucleotide sequencing.
2.3 3D modeling of protein structure
The 3D protein structure model of AMOTL1 (NM_130847.3) was predicted by I-TASSER (Roy, Kucukural, & Zhang, 2010; Yang et al., 2015; Zhang, 2008). The side chain orientation of the mutant residue was obtained by the PyMol Molecular Graphics System, v.1.5 Schrodinger, LLC. Amino acid conservation scores were calculated by Consurf at its default settings (Ashkenazy et al., 2016), based on comparison of the AMOTL1 protein sequence with homologous sequences retrieved from the UniRef90 database.
3 RESULTS
3.1 Clinical report
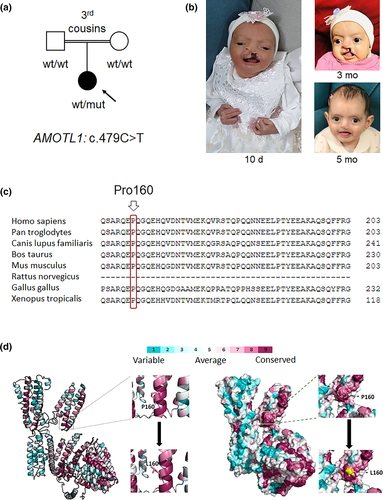
We report a seven-month-old female patient (Figure 1a,b, Table 1), first child to healthy consanguineous parents of Arab Muslim descent, in whom prenatal sonography demonstrated presence of cleft lip and palate. Fetal echocardiogram was unremarkable. Prenatal chromosomal microarray analysis (CMA) was normal. The patient was born at 37 weeks of gestation; birth weight was 3,004 g (64th %tile) and head circumference was 33.2 cm (30th %tile). Physical examination was notable for hypotonia, cleft lip and palate, imperforate anus with a recto-vaginal fistula, pilonidal dimple and dysmorphic features which included hypertelorism, micrognathia, large prominent ears with small folded earlobes and an abnormal left anti-helix, elongated fingers and hallux varus on the right foot (Figure 1b). Evaluation for associated defects demonstrated additional pathologic findings: small right-posterior choroid plexus cysts on head ultrasound (US), and a suspected tethered cord on spinal US. Renal US was normal. Postnatal echocardiography showed normal cardiac function without evidence of significant structural abnormalities. Postnatal hearing screening and ophthalmologic examination were unremarkable. The combination of findings raised suspicion of an underlying syndrome. Since prenatal CMA was not diagnostic, trio whole exome sequencing of the patient's DNA and her parents was performed. At 2 months follow-up in genetic clinic the parents reported that the girl's development was age-appropriate including presence of social smile and eye-tracking. Her weight gain was also satisfactory. At 4 months, the girl underwent successful repair of the cleft lip. By age 6 months, she was sitting with assistance and reached for objects; overall, her development was appropriate for age. Presently, she is still in need for multidisciplinary follow-up including neurosurgical assessment for suspected tethered cord, general pediatric surgery due to anal atresia with fistula and plastic surgery due to yet unrepaired cleft palate.
| Clinical features | Index proband | Liegel et al. (2019)—Proband | Liegel et al. (2019)—Affected father |
|---|---|---|---|
| Congenital cleft lip and palate | Present | Present | Present |
| Congenital cardiac malformation | Excluded | Tetralogy of Fallot, double orifice mitral valve | Atrial septal defect, reported mitral valve anomaly |
| Central nervous system anomaly | Small right posterior choroid plexus cysts | Large cisterna magna | Unknown |
| Spinal cord anomaly | Suspected tethered cord | Not mentioned | Not mentioned |
| Gastrointestinal defects | Imperforate anus with recto-vaginal fistula | Not mentioned | Not mentioned |
| Auricular abnormality | Large prominent ears with small folded earlobes and an abnormal left anti-helix | Large dysplastic ears | Large dysplastic ears |
| Skeletal deformity | Elongated fingers, hallux varus on the right foot | Short mandible, advanced bone age, bent fingers | Advanced bone age |
3.2 Exome sequencing identifies a de novo variant in AMOTL1
Analysis of trio exome sequencing data revealed a de novo novel missense variant in the AMOTL1 gene (chr11:94532835 [hg19]; NM_130847.3; c.479C>T; p.(Pro160Leu)), confirmed by Sanger sequencing. The specific variant is predicted to alter a highly conserved residue (GERP 5.0399, Figure 1c), has a CADD score of 23.8, and is predicted pathogenic by multiple bioinformatics algorithms (MutationTaster, SIFT, LRT, Provean). It is absent from the gnomAD database and from our in-house exome database. Interestingly, an additional homozygous deletion/insertion variant in the gene F12 (chr5:176831172; NM_000505.3; c.971_1018+24del) was detected. This specific variant was previously linked to autosomal dominant hereditary angioedema with partial penetrance (de Maat et al., 2016). The latter was considered a secondary finding unrelated to the primary referral diagnosis; notably, no episodes of bleeding or angioedema were documented in the medical file of the patient, and both parents were reportedly healthy without angioedema episodes.
3.3 3D structural modeling supports variant pathogenicity
In order to better predict whether the p.(Pro160Leu) variant altered relatively conserved and functional residues, in silico modeling was utilized. The p.(Pro160Leu) variant affects a highly conserved site, and results in replacement of the aromatic ring of proline by the side chain of leucine, which protrudes from the helical secondary structure (Figure 1d).
4 DISCUSSION
AMOTs, in particular, AMOTL1, are involved in regulation of organogenesis. Similar to the family reported by Liegel et al. (2019), the individual described herein harbors a heterozygous missense variant in AMOTL1 that is predicted to alter a highly conserved site. The proband suffered from oro-palatine and earlobe maldevelopment. In addition, she had abnormally long fingers and midline defects, specifically, imperforate anus and tethered cord that required multidisciplinary gradual surgical repair. The girl's intellect appeared to be unaffected. The shared phenotypic features between our patient and the family described by Liegel et al. (2019) suggest that both AMOTL1 variants are indeed pathogenic, and that pathogenic variations in human AMOTs may lead to defective organ formation such as cleft lip and palate. The phenotypic differences that are noted between the father and daughter reported by Liegel et al. and between our patient indicate variable expressivity and suggest that the full clinical spectrum associated with pathogenic variants in AMOTL1 may include additional, yet unreported malformations. Interestingly, our patient suffers from anorectal, spinal cord and limb anomalies and the previous family had cardiac defects, thereby suggesting a VACTERL-like clinical spectrum (Table 1).
Notably, the missense variant reported in Liegel et al. (2019) affects a residue only three amino acids away from that described here (p. (Arg157Cys) and p.(Pro160Leu), respectively), suggesting that there may be an important functional domain in this region. Although studies in mice with a missense Arg157Cys AMOTL1 variant were shown to result in increased embryonal death rates in both heterozygous and homozygous forms, none of the phenotypic features seen in humans with similar mutations were observed. Liegel et al. suggested that humans represented the milder effect of such variants, and that the pathology may result from a neomorphic allele acting in a dominant negative fashion (Liegel et al., 2019). Considering the low pLI score of the gene (pLI = 0) and the o/e ratio (0.3), haploinsufficiency is not a likely mechanism (Karczewski et al., 2020), urging caution of AMOTL1 variant interpretation. This seems consistent with previous studies suggesting functional redundancy between the AMOT proteins (Li et al., 2012). Thus, for the time being, the precise pathophysiologic mechanism by which pathogenic variants in AMOTL1 result in the specific phenotypes remains obscure. Provided the prominent role of Wnt/β-catenin signaling and Hippo signaling in organogenesis and specifically, in craniofacial and cardiac development (Reynolds et al., 2019; Shu, Shu, & Cheng, 2019; Zhu et al., 2019), future studies focusing on the effect of AMOTL1 pathogenic variants on these pathways may shed light as to the pathophysiologic mechanism.
In conclusion, our findings support the previous report by Liegel et al. that linked pathogenic variants in AMOTL1 in humans to a phenotype of cleft lip and palate and incompletely penetrant defects in organogenesis of various systems. Combining the findings of cardiac malformation, anal atresia and tethered cord raises the possibility that other VACTERL-like malformations may be encountered in association with AMOTL1 in the future, prompting suitable work-up. In consistency with our report, we recommend adding AMOTL1 to genetic screening panels for cleft lip and palate.
ACKNOWLEDGMENTS
The authors wish to thank the family for their participation in this study.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Jonathan Rips, Hagar Mor-Shaked, and Tamar Harel designed the study, analyzed the data, and wrote the article with input from all authors. Jonathan Rips, Smadar Eventov-Friedman, and Tamar Harel contributed clinical information, Serkan Erdin did the 3D protein modeling, and Shira Yanovsky-Dagan conducted molecular studies.
ETHICS
Informed consent was obtained from the family, in accordance with institutional review board (IRB) guidelines. Written consent was provided for publication of photographs.
Open Research
DATA AVAILABILITY STATEMENT
The ClinVar accession number for the DNA variant data is SCV001364376.




