A heterozygous, intragenic deletion of CNOT2 recapitulates the phenotype of 12q15 deletion syndrome
Funding information: Wellcome Trust.
Abstract
Only a few individuals with 12q15 deletion have been described, presenting with a disorder characterized by learning disability, developmental delay, nasal speech, and hypothyroidism. The smallest region of overlap for this syndrome was included in a genomic segment spanning CNOT2, KCNMB4, and PTPRB genes. We report on an additional patient harboring a 12q15 microdeletion encompassing only part of CNOT2 gene, presenting with a spectrum of clinical features overlapping the 12q15 deletion syndrome phenotype. We propose CNOT2 as the phenocritical gene for 12q15 deletion syndrome and its haploinsufficiency being associated with an autosomal dominant disorder, presenting with developmental delay, hypotonia, feeding problems, learning difficulties, nasal speech, skeletal anomalies, and facial dysmorphisms.
1 INTRODUCTION
Interstitial deletions of chromosome 12q are rare, only a few patients having been described so far presenting with learning disability, developmental delay, nasal speech, and hypothyroidism as common clinical features. Klein et al. (2005) suggested a new genotype–phenotype correlation, by comparing the partially overlapping deletions of three unrelated patients, presenting with growth retardation, heart disease, genitourinary anomalies, hyperkeratosis pilaris, developmental delay, and dysmorphic features. Standard and molecular-cytogenetic techniques disclosed a common deleted region spanning from 12q15-12q21.1 to 12q21.33. Schluth et al. (2008) reported a 7.7–10 Mb deletion in 12q15q21.2 in a patient presenting with clinical features superimposable to those described in the first patients, while Vergult et al. (2011), based on the genomic analysis of three other subjects, further refined the smallest region of overlap (SRO) at 12q15. This region spans 1.34 Mb of genomic DNA and includes 6 RefSeq genes, CNOT2, KCNMB4, PTPRB, PTPRR, TSPAN8, and LGR5, which have been considered responsible for a disorder characterized by learning disability, developmental delay, nasal speech, and hypothyroidism. We recently described another patient, and narrowed down the SRO to a region of 742 kb, including CNOT2, KCNMB4, and PTPRB genes (Alesi et al., 2017).
Here, we report a boy heterozygous for a 12q15 deletion involving only part of CNOT2 gene. This observation provides insight into the consequence of CNOT2 haploinsufficiency and suggests the existence of a distinct mendelian disorder overlapping in part del12q15 syndrome.
2 CLINICAL REPORT
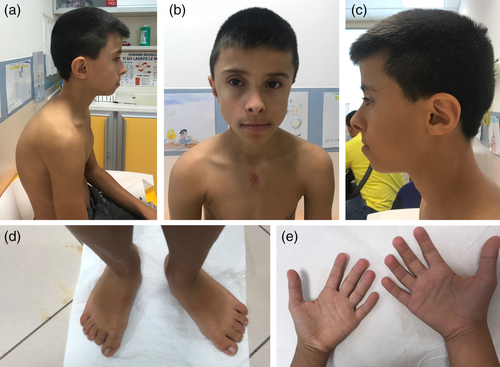
The proband is the only son of healthy nonconsanguineous parents of Romanian ancestry. Maternal and paternal age at delivery was 21 and 25 years, respectively. Prenatal period was complicated by maternal hypertension in the third trimester. The baby was born at term, with a birth weight 2.4 kg (third centile) and length 49 cm (32nd centile). Valvular and supravalvular pulmonary stenosis and mild aortic insufficiency were diagnosed at 6 months. Hypotonia and failure to thrive were recorded in the first 3 years of life. Developmental milestones were grossly delayed. He attends schooling with a specific program for children with learning difficulties. At age of 13 years, he was admitted to our Hospital for the correction of heart defect, which was complicated by delayed wound healing (>6 months) without any sign of active infection. He was diagnosed with chronic renal failure. Renal ultrasound showed increased parenchymal hyperechogenicity and bilateral absent cortico-medullary differentiation. Left kidney showed a 17 mm upper lobe cyst. Mild right renal pelvic dilatation was also present. Urine analysis disclosed albuminuria. Thyroid function test and eye tests were all unremarkable. Physical examination at age of 13 years (Figure 1) disclosed weight 34.20 kg (<3 percentile), height 146.50 cm (3–25 percentile), and occipitofrontal circumference 54 cm (25–50 percentile). The face was elongated and slightly asymmetric, with deep-set eyes and upward-slanted palpebral fissures, hypotelorism, triangular nose with anteverted nostrils, long philtrum, underbite, high-arched palate, unilateral supernumerary nipple, stubby fingers with fleshy pads at the fingers tip, flat feet with short second to fifth toes, pseudohypertrophy of calves, kyphosis, and scoliosis. Speech was remarkably nasal, and the character was friendly. The main clinical features are summarized in Table 1.
| Schluth et al. (2008) | Vergult et al. (1) (2011) | Vergult et al. (2) (2011) | Vergult et al. (3) (2011) | Lopez et al. (2012) | Alesi et al. (2017) | Our patient | |
|---|---|---|---|---|---|---|---|
| Gender | F | M | F | F | F | M | M |
| Cytogenetic position | 12q15q21.2 | 12q15q21.1 | 12q15q21.1 | 12q15q21.1 | 12q15q21.1 | 12q15 | 12q15 |
| Breakpoints (hg19) | 68,582,752–78,553,987 | 69,354,696–71,853,018 | 70,496,651–73,086,047 | 70,515,973–73,086,047 | 70,178,509–74,883,975 | 70,232,655–70,974,979 | 70,672,317–70,757,341 |
| Size of deletion | 10.21 Mb | 2.50 Mb | 2.59 Mb | 2.57 Mb | 4.71 Mb | 0.74 Mb | 0.085 Mb |
| Inheritance | De novo | De novo | De novo | De novo | De novo | De novo | De novo |
| Pregnancy and delivery | IUGR, uneventful delivery | Uneventful | NR | Uneventful | Uneventful | Uneventful | Maternal hypertension in 3rd trimester |
| Weight (kg) | 2.1 (P3) | 2.63 (P25–50) | 2.85 (P10) | 3.5 (P50) | 3.51 | 3.27 | 2.4 |
| Length | 45 (P3) | 49 | NR | NR | 49.5 | 51 | 49 |
| Head circumference | 32 | 33 (P75) | NR | NR | 37.5 | 34 | NA |
| Apgar | NR | 1 at 1 min, 6 at 5 min, 8 at 10 min | NR | NR | NR | 9 at 1 min | 8 at 5 min |
| Age at last evaluation | 5 | 16 | 11 | 21 | 4 | 29 | 13 |
| Weight at last evaluation | NR | 56.8 (P10–25) | NR | NR | −2.5 SD | NA | <3 P3 |
| Height at last evaluation | NR | 175 (P25–50) | P9 | NR | −2.5 SD | NA | P3–25 |
| OFD at last evaluation | NR | 54 (P25) | 56.5 (P98) | NR | +1.5 SD | NA | 54 (P25–50) |
| Developmental delay | + | + | + | + | + | + | Learning difficulties |
| Hypotonia | + | + | − | − | NR | + | + |
| Walking delay | + | + | − | − | + | + | NR |
| Speech delay | + | + | − | − | + | + | + |
| Feeding problems | Gastro-esophageal reflux | + | + | − | + | + | + |
| Hypothyroidism | − | + | + | Maternal in pregnancy | − | − | − |
| Heart disease | Ventricular septal defect | NR | NR | NR | − | − | Valvular and supravalvular pulmonary stenosis, mild aortic insufficiency |
| Kidney disease | NR | NR | NR | NR | − | − | Renal dysplasia and chronic renal failure |
| Genital anomalies | NR | NR | NR | NR | NR | Cryptorchidism | − |
| Skeletal anomalies | Delayed bone age, sacrococcygeal anomalies | Straight back, cubitus valgus, mild legs asymmetry, mild scoliosis, and S-shaped configuration of tibiae | NR | NR | Delayed bone age, slightly shortened clavicles | Narrow shoulders, asymmetry of pelvis and lower limbs, scoliosis, flat feet, and right valgus knee | Kyphosis, scoliosis, and flat feet |
| Microcephaly | NR | NR | + | NR | + | − | − |
| Flat face/midfacial hypoplasia | − | + | + | + | NR | + | +/− |
| Triangular-shaped face | + | − | − | NR | + | + | + |
| Large anterior fontanel | + | + | − | NR | NR | + | NR |
| High forehead | + | + | + | + | + | + | − |
| Deep-set eyes | NR | NR | NR | NR | + | + | + |
| Hyper/hypo-telorism | Hyper | Hypo | Normal | Normal | NR | Hypo | Hypo |
| Straight eyebrows | − | + | + | + | + | + | − |
| Low-set ears | + | − | + | + | NR | − | − |
| Nose shape | Linear, anteverted nostrils | NR | NR | NR | Bulbous | NR | Triangular, anteverted nostrils |
| Nasal bridge | NR | NR | NR | NR | Broad | High | Flat, narrow |
| Columella | Elongated | NR | NR | NR | Wide | − | − |
| Mouth and palate | Micrognathia | Micrognathia, narrow high palate | − | − | − | Micrognathia, oligodontia | High-arched palate |
| Long philtrum | + | + | − | − | + | + | − |
| Prognathism | NR | NR | NR | NR | NR | − | − |
| Malocclusion | NR | NR | NR | NR | NR | + | Underbite |
| Other extremities anomalies | Second to third toes syndactyly, brachydactylous fingers | Hammer toes, long slender fingers, clinodactilous fifth fingers | Hammer toes, hypoplastic toenails | NR | Clinodactyly 5th fingers | NR | Stubby fingers with pads, short second to fifth toes |
| Nasal speech | − | + | + | + | + | + | + |
| Vision impairment | NR | NR | Refractive error | Refractive error | NR | NA | − |
| Recurrent infections | NR | + | Lymphoblastic leukemia | + | NR | + | − |
| Hirsutism | Sacrococcygeal tuft of hair | Synophria | NR | NR | Mild | − | − |
| Miscellaneous | Strabismus, mild pectus excavatum | Otis media | Dyslexia, dyspraxia | Anorexia nervosa | Large thorax with widely spaced nipples | Neurosensorial hearing loss, left megaureter primary obstruction, upper limbs postural tremor, hyperelastic skin, and mild ligaments laxity | Pseudo-hypertrophy of calf muscles, unilateral supernumerary nipple |
- Abbreviations: NA, not available; NR, not reported; IUGR, intrauterine growth restriction; OFD, Occipito-Frontal Diameter.
3 MATERIALS AND METHODS
Genomic DNA was extracted from peripheral blood with Qiagen columns (QIAamp DNA minikit; Qiagen, Hilden, Germany). Chromosomal microarray analysis (CMA) was performed using Infinium CytoSNP-850K BeadChip (Illumina, San Diego, CA), according to the manufacturer's protocol. Array scanning data were generated on the Illumina NextSeq 550 system and the results were analyzed by the Bluefuse Multi 4.4 software.
Confirmation and segregation tests on patient's and parents' DNA were performed by real-time PCR (polymerase chain reaction) on CNOT2, using a SYBR Green assay (Livak & Schmittgen, 2001).
4 RESULTS AND DISCUSSION
CMA analysis detected a de novo microdeletion at 12q15, 85 kb in size, involving part of CNOT2 gene (exon 3 through exon 15). Interstitial deletions of chromosome 12q are rare, resulting in variable phenotypes depending on size and gene content (Figure 2). The reported deletions range in size from 25.5 Mb (Klein et al., 2005) to 742 kb (Alesi et al., 2017). Although the critical SRO has been restricted to 12q15 (Alesi et al., 2017; Vergult et al., 2011), the exact contribution of candidate genes to the phenotype has not been yet established.
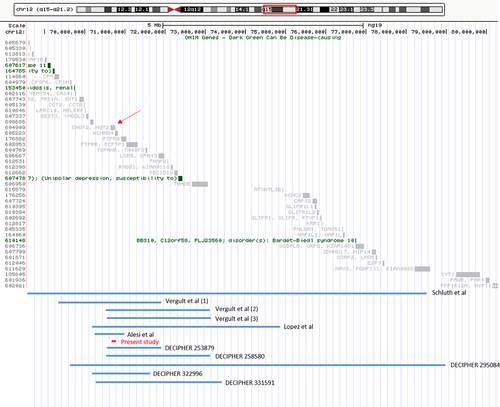
The present patient with a heterozygous deletion of a large portion of CNOT2 gene manifests overlapping features with other patients affected by deletions spanning the 12q15 region. This suggests that the critical gene for del12q15 syndrome is CNOT2. Moreover, ExAC browser (http://exac.broadinstitute.org/) states a pLI (probability of loss-of-function intolerance) score of 1.00, indicating loss-of-function intolerance of CNOT2 (Lek et al., 2016). This further corroborates the causative role of CNOT2 haploinsufficiency, which may results in a new autosomal dominant disorder.
CNOT2 encodes a subunit of the multi-component CCR4-NOT complex, involved in gene expression regulating mRNA synthesis, degradation, splicing, transport, and localization (Jeong, Kwon, Jeong, Sohn, & Kim, 2017; Rambout et al., 2016). According to its wide biological functions, it is ubiquitously expressed (https://www.genecards.org), including the nucleus, cytosol, and plasma membrane.
Table 1 summarizes the clinical features of published del12q15 patients and corroborates the role of CNOT2 haploinsufficiency in this deletion syndrome, including growth delay, feeding problems, hypotonia, learning difficulties, delayed nasal speech, high/cleft palate, and mildly abnormal extremities. Although skeletal anomalies are not cited in the majority of published patients, they could be part of the 12q15 deletion phenotype, including kyphosis, scoliosis, leg asymmetry, flat feet, and delayed bone age. Congenital heart defects have been reported in a few patients and could be a feature of CNOT2 haploinsufficiency. Presently, renal dysplasia and chronic renal failure is reported only in our patient and the association with the deleted region remains unclear. Vision impairment and hypothyroidism, described in several patients harboring larger deletions, were lacking in our patient, suggesting that these characteristics may be secondary to haploinsufficiency of other genes within the SRO. However, basing on the clinical features of the small number of patients described so far, a fine characterization of this new syndrome is still challenging. Further studies are needed for expanding the phenotypic spectrum of 12q15 deletion, leading to a better genotype–phenotype correlation.
5 CONCLUSIONS
The present patient reassesses the SRO of 12q15 deletion syndrome and identifies CNOT2 as a dose sensitive gene, probably critical for most of clinical features presented by patients harboring this deletion. Our results suggest that CNOT2 is a novel disease gene, associated with an autosomal dominant disease characterized by growth retardation, hypotonia, facial dysmorphisms, feeding problems, learning difficulties, nasal speech, and skeletal anomalies. Further observations of CNOT2 gene deletion or inactivating point mutations will improve the delineation of this novel disorder.
ACKNOWLEDGMENTS
The authors thank the child and family for their active collaboration and willingness to share the results. This project is part of Good Clinical Practice project of Bambino Gesù Children Hospital and Research Institute, “cardiogenetic approach in children with heart diseases”. The molecular characterization of this study was supported by “Ricerca Corrente 2018”: clinical and genetic analytic pathway of patients affected by rare cardiac diseases. The authors thank Technogenetics for making available NextSeq 550 (Illumina) instrument. This study makes use of data generated by the DECIPHER community. A full list of centers that contributed to the generation of the data is available from http://decipher.sanger.ac.uk and via email from [email protected]. Funding for the project was provided by the Wellcome Trust.
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.




