Noonan syndrome in diverse populations
Abstract
Noonan syndrome (NS) is a common genetic syndrome associated with gain of function variants in genes in the Ras/MAPK pathway. The phenotype of NS has been well characterized in populations of European descent with less attention given to other groups. In this study, individuals from diverse populations with NS were evaluated clinically and by facial analysis technology. Clinical data and images from 125 individuals with NS were obtained from 20 countries with an average age of 8 years and female composition of 46%. Individuals were grouped into categories of African descent (African), Asian, Latin American, and additional/other. Across these different population groups, NS was phenotypically similar with only 2 of 21 clinical elements showing a statistically significant difference. The most common clinical characteristics found in all population groups included widely spaced eyes and low-set ears in 80% or greater of participants, short stature in more than 70%, and pulmonary stenosis in roughly half of study individuals. Using facial analysis technology, we compared 161 Caucasian, African, Asian, and Latin American individuals with NS with 161 gender and age matched controls and found that sensitivity was equal to or greater than 94% for all groups, and specificity was equal to or greater than 90%. In summary, we present consistent clinical findings from global populations with NS and additionally demonstrate how facial analysis technology can support clinicians in making accurate NS diagnoses. This work will assist in earlier detection and in increasing recognition of NS throughout the world.
1 INTRODUCTION
Noonan syndrome (NS) is characterized by congenital heart disease, short stature, distinctive facial features, chest deformities, variable developmental delay, and other anomalies (Bhambhani & Muenke, 2014; Noonan, 1968). Diagnostic criteria have been established (van der Burgt et al., 1994) as well as management guidelines (Roberts, Allanson, Tartaglia, & Gelb, 2013; Romano et al., 2010). The typical facial features of NS include widely spaced eyes, down slanted palpebral fissures, ptosis, and low-set ears. The prevalence of NS is roughly 1:1,000 to 1:2,500 and is inherited in an autosomal dominant manner (Romano et al., 2010). Although NS is a common genetic syndrome, there are few phenotype and genotype studies in non-European cohorts.
The genetic etiologies of NS occur in genes associated with the Ras-mitogen-activated protein kinase (Ras/MAPK) pathway. Genes in this pathway are involved in cell differentiation, growth, and death. Other syndromes associated with Ras/MAPK genes include Costello syndrome, Cranio-facio-cutaneous (CFC) syndrome, NS with multiple lentigines (formerly called LEOPARD syndrome), and neurofibromatosis. Significant phenotypic overlap exists with other RASopathies including CFC syndrome and Costello syndrome. The most common genetic cause of NS is gain-of function mutations in the PTPN11 gene encoding the Src homology protein-tyrosine phosphatase-2 (SHP-2) with a variant occurring in 50% of individuals with NS (Tartaglia et al., 2001, 2002). PTPN11 was the first gene to be associated with NS and now there are more than eight known genes (PTPN11, SOS1, RAF1, RIT1, KRAS, NRAS, BRAF, MAP2K1, RRAS, RASA2, A2ML1, SOS2, LZTR1) that cause NS (Allanson & Roberts, 1993). SOS1 is the second most common causative gene with variants occurring in 16–20% of individuals without PTPN11 variants (Roberts et al., 2007; Tartaglia et al., 2007).
A number of investigators have evaluated PTPN11 variants in diverse populations. In a Brazilian cohort of 50 individuals with NS, 42% were found to have PTPN11 variants, but the most common variant, p.N308D, which was found in 31% of a North American cohort (Tartaglia et al., 2002) was not present (Bertola et al., 2006), demonstrating that different variants are found in different populations. In contrast, Lee et al. (2007) found half of a Korean cohort of individuals with NS to have previously reported PTPN11 variants including 29% of the cohort with the p.N308D variant (Lee, Ki, & Lee, 2007). In another Korean cohort of 59 patients, Ko et al. (2008) found that in 13 patients with PTPN11 variants, 12 had been previously reported. Although genotypes have been compared, the phenotype in NS has not been contrasted between different countries and populations. However, investigators in some countries have defined regional height phenotypes in NS and have developed growth charts specific to their country including Japan (Isojima et al., 2016) and Brazil (Malaquias et al., 2012).
NS can be a difficult diagnosis to make as the phenotype is variable and changes with age (Allanson, Hall, Hughes, Preus, & Witt, 1985; van der Burgt et al., 1999). Comprehensive characterization of NS in diverse populations has not yet been done in the medical literature. In this report, we use images, subjective examination data, and facial analysis technology to describe NS in diverse populations.
2 METHODS
2.1 Review of medical literature
Studies that characterize NS in diverse populations were obtained from a Medline search. The search terms used included: NS, Africa, Asia, Latin America, Middle East, diverse populations, and facial analysis technology. Further studies were found using reference lists of journal studies. After obtaining journal permissions, photos of individuals with NS were used to supplement study participants described below (Addissie et al., 2015; Aoki et al., 2013; Edwards et al., 2014; Lee & Sakhalkar, 2014; Ndiaye et al., 2014; Yaoita et al., 2016).
2.2 Patients
Individuals with NS were evaluated from 20 countries. All participants (Supplementary Table S1) had NS diagnosed by both clinical evaluation and/or molecular diagnosis. The patients were grouped by geographic area of origin or ethnicity (African and African American, Asian, Latin American, and Additional). Local clinical geneticists examined patients for established clinical features found in NS (van der Burgt et al., 1994).
Consent was obtained by local institutional review boards and the Personalized Genomics protocol at the National Institutes of Health (11-HG-0093). Exam findings from the current study and those from the medical literature are recorded in Table 1.
| Present study | Hung et al. (2007) | Ndiaye et al. (2014) | Bertola et al. (2006) | Yoshida et al. (2004) | Lee et al. (2007) | Ko et al. (2008) | Simsek-Kiper et al. (2013) | Essawi et al. (2013) | Jongmans et al. (2005) | Lee et al. (2011) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Africa, n = 30 | Asia, n = 36 | Latin America, n = 33 | p-values | Taiwan, n = 34 | Senegal, n = 6 | Brazil, n = 50 | Japan, n = 45 | Korea, n = 14 | Korea, n = 59 | Turkey, n = 26 | Egypt, n = 21 | The Netherlands, n = 56 | Korea, n = 59 | |
| Average age (years) | 9.3 | 8.6 | 9.9 | 9.06 | 12 | 10 | 14.8 | 8.8 | 3.7 | |||||
| Age range (years) | 0.33–30 | 0.17–31 | 0.17–31 | 1–31 | 0.3–24.1 | 0.1–34.5 | 2–29 | 0.1–17.2 | 0.25–29 | 2–20 | ||||
| PTPN11 (%) | 6/9 (67) | 22/26 (85) | 17/21 (81) | 13 (38) | 2 (33) | 21 (42) | 18 (40) | 7 (50) | 16 (27) | 7 (27) | 56 (100) | 23 (39) | ||
| SOS1 (%) | 2/26 (8) | 2/21 (10) | 10 (17) | 5 (19) | 12 (20) | |||||||||
| SHOC2 (%) | 0 | 3 (12) | ||||||||||||
| Widely spaced eyes | 80% | 96% | 94% | 0.10 | 6 (100%) | 22 (44%) | 45 (100%) | 22 (85%) | 21 (100%) | |||||
| Ptosis | 63% | 72% | 94% | 0.011 | 12 (35%) | 25 (96%) | 6 (29%) | |||||||
| Downslanted palpebral fissures | 87% | 86% | 73% | 0.25 | 20 (59%) | 33 (66%) | 19 (73%) | 21 (100%) | ||||||
| Epicanthal folds | 70% | 64% | 55% | 0.44 | 19 (56%) | |||||||||
| Low-set ears | 82% | 94% | 88% | 0.30 | 1 (17%) | 15 (58%) | 12 (57%) | |||||||
| Lowset posterior hairline | 64% | 76% | 69% | 0.57 | 25 (74%) | |||||||||
| Webbed neck | 57% | 36% | 69% | 0.023 | 21 (62%) | 3 (50%) | 46 (92%) | 10 (71%) | 16 (62%) | 21 (100%) | 10 (18%) | 48.30% | ||
| Pulmonary stenosis | 50% | 53% | 48% | 0.94 | 12 (35%) | 2 (33%) | 28 (56%) | 16 (36%) | 8 (57%) | 24 (41%) | 16 (62%) | 5 (24%) | 38(68%) | 22/43 (51%) |
| Hypertrophic cardiomyopathy | 7% | 11% | 10% | 0.82 | 3 (50%) | 6 (12%) | 5 (11%) | 13 (22%) | 2 (8%) | 4 (19%) | 4 (7%) | 11/43 (26%) | ||
| ASD | 27% | 14% | 24% | 0.39 | 14 (31%) | 6 (43%) | 19 (32%) | 11 (42%) | 17 (30%) | 12/43 (28%) | ||||
| VSD | 7% | 17% | 6% | 0.26 | 2 (14%) | 13 (22%) | 3 (12%) | 7/43 (16%) | ||||||
| Septal defects | 37% | 36% | 27% | 0.66 | 16 (47%) | 11 (79%) | 14/26 (54%) | |||||||
| PDA (%) | 0 | 8 | 0 | 7 (12) | 7/43 (16) | |||||||||
| Short stature (<3rd centile%) | 71% | 80% | 83% | 0.53 | 32 (94%) | 6 (100%) | 48 (96%) | 9 (64%) | 30 (51%) | 14/26 (54%) | 15 (71%) | 41 (73%) | 27/41 (66%) | |
| Chest deformity | 59% | 49% | 70% | 0.21 | 13 (38%) | 4 (67%) | 28 (56%) | 9 (20%) | 9 (64%) | 14 (24%) | 17 (81%) | 24 (43%) | 16/43 (37%) | |
| Scoliosis | 37% | 17% | 13% | 0.08 | 9/47 (19%) | |||||||||
| Undescended testes | 47% | 26% | 47% | 0.17 | 1/2 (50%) | 14/27 (52%) | 9/25 (36%) | 4/9 (44%) | 19/41 (46%) | 4/12 (33%) | 27/32 (84%) | 11/29 (38%) | ||
| Coagulopathy | 26% | 14% | 6% | 0.12 | 20/48 (42%) | 5 (11%) | 3 (5.1%) | 32 (57%) | 4/43 (9%) | |||||
| Learning disability/intellectual disability | 64% | 54% | 63% | 0.69 | 19 (56%) | 11/43 (26%) | 12 (46%) | 11 (52%) | 28 (50%) | 11/40 (28%) | ||||
| Renal anomalies | 11% | 17% | 10% | 0.75 | 7/50 (14%) | |||||||||
| Skin pigmentation lesions | 12% | 38% | 27% | 0.12 | ||||||||||
2.3 Facial analysis technology
As previously described (Kruszka, Addissie, et al., 2017; Kruszka, Porras, et al., 2017b), digital facial analysis technology (Cerrolaza et al., 2016; Zhao et al., 2013; Zhao, Okada, et al., 2014; Zhao, Werghi et al., 2014) was used to evaluate 161 images of individuals with NS, and 161 ethnic, gender, and age matched controls from our previously described database (Zhao, Okada, et al., 2014; Zhao et al., 2013; Zhao, Werghi, et al., 2014). The 161 individuals with NS used for facial analysis technology included individuals from Supplementary Table S1 and additional archival images of individuals with NS. A Caucasian ethnic group was identified in addition to African, Asian, and Latin American for the purpose of facial analysis. The distribution of the dataset is presented in Table 2. Only frontal images were analyzed by this technology.
| Noonan syndrome (N = 161) | Controls (N = 161) | |||
|---|---|---|---|---|
| Age | Number | % | Number | % |
| Newborn | 0 | 0 | 0 | 0 |
| Infant | 45 | 28 | 45 | 28 |
| Toddler | 29 | 18 | 29 | 18 |
| Child | 47 | 29 | 47 | 29 |
| Adolescence | 18 | 11 | 18 | 11 |
| Adult | 22 | 14 | 22 | 14 |
| Total | 161 | 161 | ||
| Ethnicity | ||||
| African Descent | 35 | 22 | 35 | 22 |
| Asian | 40 | 25 | 40 | 25 |
| Caucasian | 40 | 25 | 40 | 25 |
| Latino | 46 | 29 | 46 | 29 |
| Total | 161 | 161 | ||
| Gender | ||||
| Male | 93 | 58 | 93 | 58 |
| Female | 68 | 42 | 68 | 42 |
| Total | 161 | 161 | ||
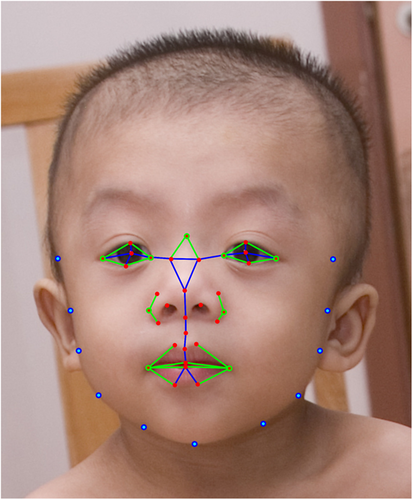
Our algorithms analyzed the images of our study participants with output variables consisting of feature extraction, feature selection, and classification. As in our previous studies (Kruszka, Addissie, et al., 2017; Kruszka, Porras, et al., 2017b), from a set of 44 landmarks placed on the frontal face images, a total of 126 facial features, including both geometric and texture biomarkers, were isolated. The geometric biomarkers consisted of a set of distances and angles calculated between the different inner facial landmarks. Figure 1 represents both the landmark locations and the geometric features extracted. Texture patterns (Cerrolaza et al., 2016) were calculated at each of the acial landmarks to quantify texture information (Figure 1). From the collection of geometric and texture features, the most significant ones were selected using the method proposed previously (Cai, Zhang, & He, 2010). For each feature set, a support vector machine classifier (Cortes & Vapnik, 1995) was trained using a leave-one-out cross-validation strategy (Elisseeff & Pontil, 2003). The optimal number of features was selected as the minimum number for which the classification accuracy converged to its maximum; Supplementary Figures S1–S5 graphically demonstrate how the addition of features improves the measures of sensitivity, specificity, and accuracy. As an estimator of the individual discriminant power of each feature selected, the p-value of each feature was also estimated using the Student's t-test. Significance between methods used to detect NS was assessed using Fisher's exact test.
3 RESULTS
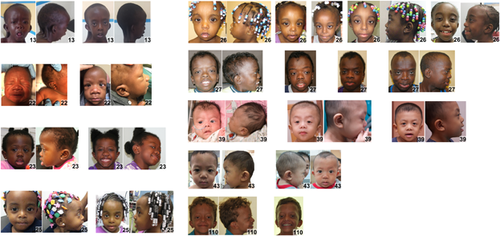
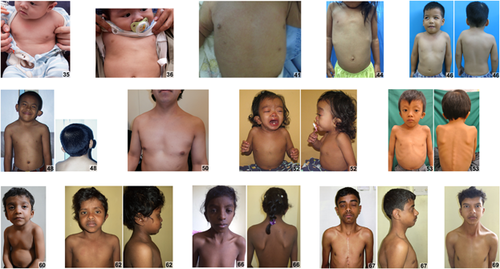
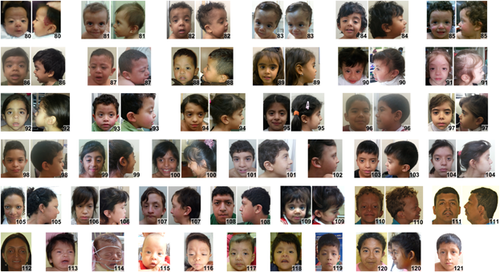
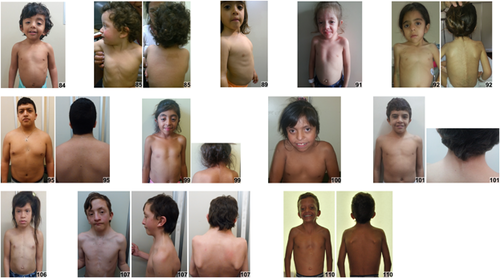
Clinical information and images were collected on 125 individuals (13 individuals were obtained from the medical literature) from 20 countries, average age was 8 years, the median age was 5 years, and 46% were females (Supplementary Table S1). Individuals of African descent are shown in Figure 2 (facial), Figure 3 (chronological sequence images), and Figure 4 (facial and torso profiles); Asian in Figure 5 (facial), Figure 3 (chronological sequence images), and Figure 6 (facial and torso profiles); Latin American in Figure 7 (facial), Figure 3 (chronological sequence images), and Figure 8 (facial and torso profiles); and additional patients in Supplementary Figure S6. Supplementary Figures S7 and S8 display hand and feet images, respectively. From the medical literature in Table 1, we found 10 non-European descent studies of NS that evaluated at least five participants and at least one facial feature (Bertola et al., 2006; Essawi et al., 2013; Hung et al., 2007; Jongmans et al., 2005; Ko, Kim, Kim, & Yoo, 2008; Lee et al., 2011; Lee et al., 2007; Ndiaye et al., 2014; Simsek-Kiper et al., 2013; Yoshida et al., 2004). We compared unpublished patients from the present study with the above-mentioned studies from the medical literature (Table 1). The most common phenotype element in both the present study and the medical literature is widely spaced eyes. In our study, all population groups had widely spaced eyes in 80% or greater of individuals, and in the medical literature, four of five studies report 85% or more of their cohorts as having widely spaced eyes (Table 1). Low-set ears, also common in our cohort, was found in over 80% of the present study but not consistently reported in the literature. And lastly, short stature as defined by <3rd centile (when centiles were provided) was found in greater than 70% of the present study and in seven of the nine studies in the medical literature. The remainder of clinical exam findings in the present study were consistent between the different population groups; the only exam elements that differed statistically among groups in the present study were ptosis and webbed neck (p = 0.01 and p = 0.02, respectively; χ2 test). Consistent with the medical literature (Allanson & Roberts, 1993), this study's most common congenital heart disease was pulmonary stenosis, found in roughly 50% of all three population groups (Table 1).



As a more objective measure of phenotype, but limited to facial features, facial analysis technology was applied to 161 individuals (Caucasian, African, Asian, and Latin American) with results shown in Table 3. The sensitivity and specificity to discriminate between NS and controls was 0.88 and 0.89, respectively, when the entire cohort was evaluated concurrently. The test accuracy of the facial recognition technology increased significantly when the cohort was analyzed by specific ethnic population (p-value < 0.001 for all comparisons), with sensitivities and specificities for Caucasian, African, Asian, and Latin American of 0.95 and 0.93, 0.94 and 0.91, 0.95 and 0.90, and 0.96 and 0.98, respectively (Table 3).
| Number of features | AUC | Accuracy | Sensitivity | Specificity | |
|---|---|---|---|---|---|
| Global | 10 | 0.94 | 0.89 | 0.88 | 0.89 |
| Caucasian | 11 | 0.98 | 0.94 | 0.95 | 0.93 |
| African and African American | 5 | 0.94 | 0.93 | 0.94 | 0.91 |
| Asian | 10 | 0.95 | 0.93 | 0.95 | 0.90 |
| Latin American | 6 | 0.97 | 0.97 | 0.96 | 0.98 |
- AUC, area under the receiver operating characteristic curve.
4 DISCUSSION
We present the first study that evaluates the clinical presentation of NS and uses facial analysis technology in diverse populations. Both clinical diagnostic guidelines (van der Burgt et al., 1994) and facial analysis technology (Hammond et al., 2004) have been reported for the diagnosis of NS cohorts in the past, but not in multiple ethnic population groups. Hammond et al. (2004) used an elaborate combination of deep surface models from three-dimensional scans combined with pattern recognition algorithms to allow for a sensitivity of 88% and specificity of 94% to discriminate between NS and controls. However, that study examined only patients of European descent and is not applicable to facial photographs.
In this study, we demonstrate that the clinical presentation of NS is similar across different population groups. When looking at 21 clinical characteristics (Table 1), only two elements where statistically different between the African, Asian, and Latin American groups: ptosis and webbed neck. Three clinical characteristics in our study were present in over 70% of participants including widely spaced eyes (≥80%), low-set ears (>80%), and short stature (>70%).
Experienced clinicians are often able to make a diagnosis of NS by recognizing characteristic facial features of NS. Allanson et al. (2010) concluded after subjective clinical exam by two well-trained and experienced clinical geneticists that facial features alone are not sufficient to predict a patient's genotype due to the presence of atypical features in some of the patients. Given the potential difficulties in clinically recognizing NS, especially when the presentation is atypical, facial analysis technology can be a useful complement to the physician's dysmorphology examination. The facial analysis technology used in our study was able to diagnose patients from all population groups with a sensitivity and specificity of 88% and 89%, respectively (Table 3). There was a significant improvement when separately evaluating population groups by the facial analysis algorithm, which led to sensitivity equal to or greater than 94%, and specificity equal or greater than 90% for all groups (Table 3). The technology identified quantitative facial biometrics specific to NS for each ethnic group. As expected, our algorithm for facial analysis found widely spaced eyes as a significant facial feature in all ethnic groups (Supplementary Tables S3–S6) as well as for the global population (Supplementary Table S2).
There are several limitations inherent to studies of genetic syndromes in diverse populations. We acknowledge that ascertainment bias exists with only the most severe phenotypes or those with severe congenital heart disease seeking medical attention. Thus, the milder cases of NS are most likely missed, as seen in adults who are often diagnosed only after their more severely affected child is diagnosed; this is further reinforced by the fact that 30–75% of individuals with NS have an affected parent (Allanson & Roberts, 1993). Additionally, in countries with limited resources and access to medical care, molecular genetic testing is difficult compared to developed countries where molecular testing is more widely available. Due to this limitation, we only accepted patients into this study who were diagnosed clinically with NS by a trained clinical geneticist since molecular genetic testing was unavailable in a fraction of our cohort (Supplementary Table S1). Another challenge to these studies is arbitrarily grouping populations geographically, for example, Chinese, Indian, and Malaysian in the category of “Asian.” Obviously, every population group is unique, and within countries a significant amount of ethnic diversity and admixture exists. As larger cohorts are assembled through public databases (Muenke, Adeyemo, & Kruszka, 2016), more precise population characterizations will be possible. Additionally, our study does not account for genotype-phenotype correlations which are known to exist, such as pulmonary valve stenosis being more common in individuals with PTPN11 variants (Tartaglia et al., 2002), or hypertrophic cardiomyopathy being more common in those with RIT1 variants (Aoki et al., 2013; Yaoita et al., 2016). However, it is important to note that Allanson et al. (2010) did not find a relationship between genotype and specific facial features in individuals with NS (Allanson et al., 2010). Finally, it is known that the facial features of individuals with NS change over time making potential genotype-phenotype correlations of this disease aspect difficult to assess (Allanson et al., 1985). Even with the above study limitations, our clinical and facial analysis data appear to be consistent and accurate in the evaluation of NS based on the available data. We would like to emphasize that facial analysis technology is a tool and not a substitute for clinical evaluation as it does not consider other important features of NS such as webbed neck, chest deformities, and congenital heart disease.
Lastly, this study and similar reports (Kruszka, Addissie, et al., 2017; Kruszka, Porras, et al., 2017b) and our recently created website, www.genome.gov/atlas will have widespread clinical significance for the diagnosis of individuals with NS, especially in countries without access to genetic services or genetic testing where the simplicity of facial analysis technology may be a useful asset.
ACKNOWLEDGMENTS
We are grateful to the individuals and their families who participated in our study. P.K., Y.A.A, and M.M. are supported by the Division of Intramural Research at the National Human Genome Research Institute, NIH. We thank the Chulalongkorn Academic Advancement into its 2nd Century Project. Partial funding of this project was from a philanthropic gift from the Government of Abu Dhabi to the Children's National Health System. We would like to acknowledge GeneDx and Dr. Benjamin Solomon for providing molecular testing for NS free of charge.