Phenotypes and genotypes in individuals with SMC1A variants
Abstract
SMC1A encodes one of the proteins of the cohesin complex. SMC1A variants are known to cause a phenotype resembling Cornelia de Lange syndrome (CdLS). Exome sequencing has allowed recognizing SMC1A variants in individuals with encephalopathy with epilepsy who do not resemble CdLS. We performed an international, interdisciplinary study on 51 individuals with SMC1A variants for physical and behavioral characteristics, and compare results to those in 67 individuals with NIPBL variants. For the Netherlands all known individuals with SMC1A variants were studied, both with and without CdLS phenotype. Individuals with SMC1A variants can resemble CdLS, but manifestations are less marked compared to individuals with NIPBL variants: growth is less disturbed, facial signs are less marked (except for periocular signs and thin upper vermillion), there are no major limb anomalies, and they have a higher level of cognitive and adaptive functioning. Self-injurious behavior is more frequent and more severe in the NIPBL group. In the Dutch group 5 of 13 individuals (all females) had a phenotype that shows a remarkable resemblance to Rett syndrome: epileptic encephalopathy, severe or profound intellectual disability, stereotypic movements, and (in some) regression. Their missense, nonsense, and frameshift mutations are evenly spread over the gene. We conclude that SMC1A variants can result in a phenotype resembling CdLS and a phenotype resembling Rett syndrome. Resemblances between the SMC1A group and the NIPBL group suggest that a disturbed cohesin function contributes to the phenotype, but differences between these groups may also be explained by other underlying mechanisms such as moonlighting of the cohesin genes.
1 INTRODUCTION
“Doctor, really wonderful that you have found that our boy has a SMC1A mutation! But please, what does that mean for him, and what can we expect?” In an era dominated by diagnostic tests using microarrays and exome sequencing that identify gene variants, this is in fact a major question that patients and their families like to be answered. This manuscript tries to provide some first answers to that question.
The first clinical reports on SMC1A described that variants in this gene cause X-linked Cornelia de Lange syndrome or a mild variant of Cornelia de Lange syndrome (CdLS) (Borck et al., 2007; Deardorff et al., 2007; Musio et al., 2006). CdLS is a multisystem disorder characterized by intrauterine growth retardation, short stature, typical face, congenital anomalies of especially the distal upper limbs, and intellectual and developmental disabilities. Behavioral characteristics include autism spectrum disorder, and a predisposition to engage with challenging behavior, especially self-injurious behavior (SIB) (Huisman et al., in press, 2017; Moss, Howlin, Magiati, & Oliver, 2012; Mulder et al., 2016; Oliver, Sloneem, Hall, & Arron, 2009). CdLS is associated with variants in a series of genes; variants in NIPBL (∼70–75%) and SMC1A (∼5%) are the most prevalent (Bhuiyan et al., 2006; Huisman, Redeker, Maas, Mannens, & Hennekam, 2013; Krantz et al., 2004; Tonkin, Wang, Lisgo, Bamshad, & Strachan, 2004).
The CdLS phenotype caused by SMC1A variants overlaps with the phenotype in individuals with NIPBL variants. Individuals with SMC1A variants were first reported with less marked facial features, less effects on growth, and without limb reduction defects (Borck et al., 2007; Deardorff et al., 2007; Musio et al., 2006). Subsequent publications have reported on a more variable phenotype (Ansari et al., 2014; Basel-Vanagaite et al., 2016; Chatfield et al., 2012; Gervasini et al., 2013; Hoppman-Chaney, Jang, Jen, Babovic-Vuksanovic, & Hodge, 2012; Limongelli et al., 2010; Liu, Feldman, et al., 2009; Liu Zhang et al., 2009; Mannini, Liu, Krantz, & Musio, 2010; Parenti et al., 2014; Pie et al., 2010, 2016; Rohatgi et al., 2010; Yuan et al., 2015). Through the use of panel screening aimed at identifying variants in genes linked to intellectual disability, and the use of untargeted trio exome analysis, SMC1A variants are increasingly detected in individuals in whom CdLS was not clinically suspected. In some of these patients the main manifestation is an epileptic encephalopathy (de Ligt et al., 2012; Fieremans et al., 2016; Gilissen et al., 2014; Goldstein et al., 2015; Hansen, Mohr, Burki, & Lemke, 2013; Jang, Lee, Kim, & Ki, 2015; Jansen et al., 2016; Lebrun et al., 2015; Tzschach et al., 2015; Wenger et al., 2016).
This urged us to initiate an interdisciplinary study in a relatively large series of individuals with a confirmed SMC1A mutation. We aimed to gather data on their physical and behavioral phenotype, and to compare the data to a series of individuals with CdLS in whom NIPBL variants were found (Bhuiyan et al., 2006; Yan et al., 2006). Here we report on the detailed results of the physical studies and on the results of the behavioral studies in general; detailed results of the behavioral studies will be published elsewhere (Mulder et al., 2016).
2 METHODS
2.1 Study design
We performed a cross-sectional study in a large international series of individuals with pathological SMC1A variants, using in person evaluations in Dutch participants, and questionnaire results and clinical pictures in patients from other countries.
2.2 Dutch SMC1A cohort
The molecular genetic laboratory of the Academic Medical Center in Amsterdam has been the central Dutch site to perform panel analysis to detect variants in any of the genes associated with CdLS, and SMC1A mutation analysis by Sanger sequencing. We contacted the physicians in charge of all individuals with pathological SMC1A variants, asking them to obtain permission for us to contact the family. Subsequently, we contacted all Dutch molecular laboratories that perform exome sequencing and asked whether they had detected additional SMC1A variants either using panel screening for intellectual disability/epilepsy or using untargeted trio analysis. Eleven families were contacted of which ten families (13 patients) agreed to participate in the study. After written consent, two authors (S.H.; R.C.H.) performed clinical evaluations (medical history, physical and morphological examination, clinical pictures) in 10 individuals and collected data from three individuals who had passed away. Two other authors (P.A.M.; A.L.) performed direct behavioral assessments (ADOS & Bayley-III-NL/WPPSI-III-NL/WAIS-IV-NL) and interviews (SSP-NL and VABS-2) in eight of the remaining individuals (one had died in the meantime; one could not be contacted for further behavioral studies). In addition, we asked parents to fill out a set of behavioral questionnaires, which included the Repetitive Behavior Questionnaire (RBQ), Challenging Behavior Questionnaire (CBQ), and Gastro-esophageal Reflux Questionnaire (GRQ).
2.3 International SMC1A cohort
We invited the members of the Scientific Advisory Committee of the CdLS World Federation from Denmark, France, Germany, Italy, Poland, Portugal, Spain, Sweden, U.K., and U.S.A. to participate, requesting to identify individuals with pathological variants in their series, and to contact their molecular genetic laboratories to check for additional SMC1A variants. We forwarded a comprehensive, dedicated questionnaire on somatic characteristics (morphology, malformations, neurodevelopment, physical health; see Supplemental materials) to the physicians and requested to forward a set of behavioral questionnaires to the families.
2.4 NIPBL comparison group
We collected data from the Polish CdLS database of individuals with NIPBL pathological variants (n = 43), some of whom were included in previous publications (Kuzniacka et al., 2013; Yan et al., 2006), and from a previously published Dutch cohort with NIPBL pathological variants (n = 24) (Bhuiyan et al., 2006). To both sets we added data that have become available since publications.
2.5 Severity score
A severity score can be predictive of clinical course and maturation relative to other individuals affected by the same or related entity. Since Gillis et al. (2004) proposed the first severity classification system based on three CdLS phenotype parameters (limb reduction, cognitive abilities, and growth), the severity scoring system has been modified and refined (Bhuiyan et al., 2006; Kline et al., 2008). We used the classification system as suggested by Bhuiyan et al. (2006), as it includes all major CdLS parameters (facial morphology, limb anomalies, growth parameters [prenatal; postnatal; skull] and cognitive/adaptive level of abilities) in a standardized and non-interdependent manner.
2.6 Statistics
Data were stored in Excel format. Descriptive statistics and Chi square test were performed using Microsoft Excel version 2011. Behavioral data were converted from the questionnaires into a coded SPSS file and were analyzed using IBM SPSS Statistics version 23.
2.7 Ethics
The present study has been supported by the national and international CdLS Support Groups, and approved by the Medical Ethics Committee of the Academic Medical Center in Amsterdam (NL39553.018.12).
3 RESULTS
We collected data from 51 individuals with pathological SMC1A variants (36 missense, 15 other types). Participants originated from the Netherlands (13 [25%]), USA (9 [18%]), the UK (8 [16%]), and smaller numbers from Argentina, Austria, Denmark, France, Germany, India, Italy, Spain, Switzerland, and Turkey. Somatic questionnaires were completed from all 51 participants. Behavioral questionnaires were obtained from 31 participants (response rate 60%). Median age was 13 years (range: 0–46 years), gender ratio was 14M to 37F. Median age of clinical diagnoses was 5 years (range: 0–46 years), median age of last examination was 11 years (range: 0–40 years). Median age of the NIPBL group was 14 years (range: 0–46 years), gender ratio was 34M to 33F.
3.1 Physical phenotypes
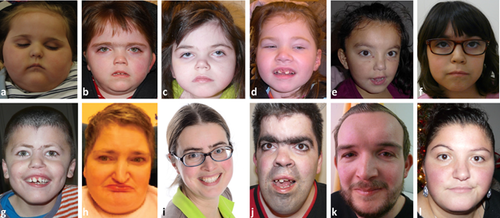
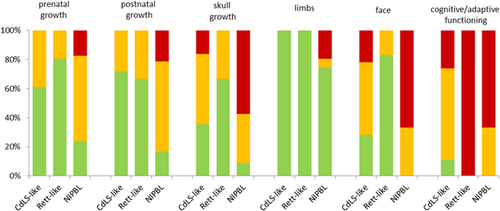
The faces of the Dutch patients are depicted in Figure 1. The main results of the present study are presented in Tables 1-4. The data in the SMC1A group are compared to the 67 individuals with NIPBL variants. The severity scores in CdLS-like, Rett-like, and NIPBL positive individuals is depicted in Figure 2. In the text we only mention those data that are not presented in the tables.
| All n = 51 | Missense variants n = 36 | Other variants n = 15 | NIPBL literature n = 67 | |
|---|---|---|---|---|
| Gender (M/F) | 14 (27)/37 (73) | 11 (31)/25 (69) | 3 (20)/12 (80) | 34 (51)/33 (49) |
| Growth | ||||
| Prenatala | ||||
| Length at birth <−2SD | 9/32 (28) | 6/21 (29) | 3/11 (27) | 32/43 (74) |
| Weight at birth <−2SD | 11/41 (27) | 8/27 (30) | 3/14 (21) | 29/43 (67) |
| Head circumference <−2SD | 8/24 (33) | 5/18 (28) | 3/6 (50) | 39/43 (91) |
| Postnatalb | ||||
| Height <−2SD | 24/38 (63) | 17/27 (63) | 7/11 (64) | 37/43 (86) |
| Weight <−2SD | 14/37 (38) | 11/26 (42) | 3/11 (27) | 39/43 (91) |
| Head circumference <−2SD | 23/36 (64) | 18/26 (69) | 5/10 (50) | 54/62 (87) |
| Craniofacial morphology | ||||
| Brachycephaly | 17/42 (40) | 12/30 (40) | 5/12 (42) | 44/67 (66) |
| Low anterior/posterior hairline | 30/43 (70) | 23/31 (74) | 7/12 (58) | 57/67 (85) |
| Arched eyebrows | 32/44 (73) | 26/31 (84) | 6/13 (46) | 54/67 (81) |
| Synophrys | 37/46 (80) | 29/33 (88) | 8/13 (62) | 61/67 (91) |
| Long eyelashes | 38/45 (84) | 27/32 (84) | 11/13 (85) | 65/67 (97) |
| Depressed nasal bridge | 20/43 (47) | 14/30 (47) | 6/13 (46) | 57/67 (85) |
| Anteverted nostrils | 26/46 (57) | 21/33 (64) | 5/13 (38) | 58/67 (87) |
| Long, featureless philtrumc | 27/43 (63) | 20/30 (67) | 7/13 (54) | 54/67 (81) |
| Thin upper vermillionc | 33/44 (75) | 26/31 (84) | 7/13 (54) | 22/24 (92) |
| Downturned corners mouth | 33/46 (72) | 24/33 (73) | 9/13 (69) | 23/24 (96) |
| Palate (high arched; cleft) | 11/37 (30); 10/45 (22) | 8/26 (31); 7/32 (22) | 3/11 (27); 3/13 (23) | 35/67 (52); 20/67 (30) |
| Widely spaced teeth | 13/44 (30) | 8/31 (26) | 5/13 (38) | 18/23 (78) |
| Micrognatia | 18/45 (40) | 16/32 (50) | 2/13 (15) | 50/67 (75) |
| Low-set and/or malformed ears | 18/45 (40) | 15/32 (47) | 3/13 (23) | 45/67 (67) |
| Limbs | ||||
| Small hands | 32/45 (71) | 23/32 (72) | 9/13 (69) | 53/63d (84) |
| Proximally placed thumb | 18/44 (41) | 13/31 (42) | 5/13 (38) | 11/20 (55) |
| Clinodactyly 5th finger | 21/45 (47) | 17/32 (53) | 4/13 (31) | 42/63 (67) |
| Syndactyly | 1/37 (3) | 1/26 (4) | 0/11 (0) | 4/63 (6) |
| Small feet | 29/44 (66) | 20/31 (65) | 9/13 (69) | 65/67 (97) |
| Syndactyly 2nd-3rd toes | 13/46 (28) | 9/33 (27) | 4/13 (31) | 21/66 (32) |
| Skin | ||||
| Cutis marmorata | 19/44 (43) | 15/32 (47) | 4/12 (33) | 27/43 (63) |
| Hirsutism | 37/47 (79) | 28/34 (82) | 9/13 (69) | 37/43 (86) |
| Major and minor malformations | ||||
| Limb (major) | 0/49 (0) | 0/35 (0) | 0/14 (0) | 17/67 (25) |
| Heart (major and minor) | 13/44 (30) | 10/32 (31) | 3/12 (25) | 18/66 (27) |
| Genitourinary system (major; minor)e | 4/42 (10); 9/40 (23) | 2/30 (7); 7/29 (24) | 2/12 (17); 2/11 (18) | 0/67 (0); 46/67 (69) |
| Gut | 3/44 (7) | 3/32 (9) | 0/12 (0) | 6/24 (25) |
| CNS | 5/43 (12) | 4/31 (13) | 1/12 (8) |
- Percentages are shown in brackets.
- Blank cell indicates that information was unavailable or uncertain.
- a In three prematurely born individuals (between 31 and 35 weeks) growth data were corrected for a gestational age of 40 weeks.
- b Postnatal data are not available in one stillborn child.
- c In three patients this could not be reliably scored due to surgery for clefting.
- d Seven of the others had such marked limb reduction defects that it prevented evaluation of hand size.
- e Major: uni/bilateral renal anomalies; minor: cryptorchidism; small penis; hypospadias; underdeveloped prepuce; small labia.
| SMC1A | NIPBL | ||||||
|---|---|---|---|---|---|---|---|
| All n = 51 | Missense variants n = 36 | Other variants n = 15 | All variants n = 67 | ||||
| Physical health | |||||||
| Apgar at 1′ <6 | 5/25 (20) | 1/14 (7) | 4/11 (36) | 18/43 (42) | |||
| Apgar at 1′ 7-10 | 20/25 (80) | 13/14 (93) | 7/11 (64) | 25/43 (58) | |||
| Apgar at 5′ <6 | 2/25 (8) | 0/14 (0) | 2/11 (18) | 11/43 (36) | |||
| Apgar at 5′ 7-10 | 23/25 (92) | 14/14 (100) | 9/11 (82) | 32/43 (74) | |||
| Feeding problems | 24/34 (71) | 17/23 (74) | 7/11 (64) | 65/67 (97) | |||
| Seizures | 20/44 (45) | 13/32 (41) | 7/12 (58) | 10/66 (15) | |||
| Gastroesophageal reflux disease | 25/42 (60) | 17/30 (57) | 8/12 (67) | 47/66 (71) | |||
| Constipation | 18/42 (43) | 14/30 (47) | 4/12 (33) | 21/66 (32) | |||
| Visual impairment | 20/38 (53) | 15/29 (52) | 5/9 (56) | 29/66 (44) | |||
| Hearing impairment | 16/39 (41) | 12/30 (40) | 4/9 (56) | 43/66 (65) | |||
| Development | |||||||
| Cognitive functioninga | |||||||
| Dutch cohortb (n = 13) | |||||||
| Normal | 1/8 (13) | 1/6 (17) | 0/2 (0) | 0/58 (0) | |||
| Mild disability | 2/8 (25) | 2/6 (33) | 0/2 (0) | 4/58 (7) | |||
| Moderate disability | 1/8 (13) | 1/6 (17) | 0/2 (0) | 16/58 (28) | |||
| Severe disability | 1/8 (13) | 1/6 (17) | 0/2 (0) | 27/58 (47) | |||
| Profound disability | 3/8 (38) | 1/6 (17) | 2/2 (0) | 11/58 (19) | |||
| International cohortc (n = 39) | |||||||
| Normal | 2/20 (10) | 1/12 (8) | 1/8 (13) | ||||
| Mild disability | 4/20 (20) | 2/12 (17) | 2/8 (25) | ||||
| Moderate disability | 8/20 (40) | 4/12 (33) | 4/8 (50) | ||||
| Severe disability | 5/20 (25) | 5/12 (42) | 0/8 (0) | ||||
| Profound disability | 1/20 (5) | 0/12 (0) | 1/8 (13) | ||||
| Sittingd | 33/38 (87) | 23/24 (96) | 10/14 (71) | 52/67 (78) | |||
| Milestone at 0–2 yrs | 19/24 (79) | 12/15 (80) | 6/9 (67) | 28/52 (54) | |||
| Milestone at 3–4 yrs | 3/24 (13) | 2/15 (13) | 1/9 (11) | 17/52 (33) | |||
| Milestone at ≥5 yrs | 0/24 (0) | 0/15 (0) | 0/9 (0) | 6/52 (12) | |||
| No milestone yet (≥5 yrs) | 3/24e (13) | 1/15e (7) | 2/9e (22) | 1/52 (2) | |||
| Walkingc | 33/39 (85) | 23/25 (92) | 9/13 (69) | 52/67 (78) | |||
| Milestone at 0–2 yrs | 17/30 (57) | 13/22 (59) | 4/8 (50) | 3/52 (6) | |||
| Milestone at 3–4 yrs | 5/30 (17) | 4/22 (18) | 1/8 (13) | 1/52 (2) | |||
| Milestone at ≥5 yrs | 4/30 (13) | 3/22 (14) | 1/8 (13) | 11/52 (21) | |||
| No milestone yet (≥5 yrs) | 4/30e (13) | 2/22e (9) | 2/8e (25) | 19/52 (37) | |||
| First wordsc | 23/35 (66) | 15/22 (68) | 7/12 (58) | 53/67 (79) | |||
| Milestone at 0–2 yrs | 7/20 (35) | 4/14 (29) | 3/6 (50) | 4/53 (8) | |||
| Milestone at 3–4 yrs | 3/20 (15) | 3/14 (21) | 0/6 (0) | 16/53 (30) | |||
| Milestone at ≥5 yrs | 1/20 (5) | 1/14 (7) | 0/6 (0) | 0/53 (0) | |||
| No milestone yet (≥ 5yrs) | 9/20e (45) | 6/14e (43) | 3/6 (50) | 33/53 (62) | |||
| Behavioral direct assessment | |||||||
| Adaptive functioning | |||||||
| Dutch cohortb (n = 13) | |||||||
| Communication | |||||||
| Mild-moderate deficit | 2/6 (33) | 2/4 (50) | 0/2 (0) | ||||
| Severe deficit | 1/6 (17) | 1/4 (25) | 0/2 (0) | ||||
| Profound deficit | 3/6 (50) | 1/4 (25) | 2/2 (100) | ||||
| Daily living skills | |||||||
| Mild deficit | 2/6 (33) | 2/4 (50) | 0/2 (0) | ||||
| Moderate-severe deficit | 1/6 (17) | 1/4 (25) | 0/2 (0) | ||||
| Profound deficit | 3/6 (50) | 1/4 (25) | 2/2 (100) | ||||
| Socialization | |||||||
| Mild deficit | 2/6 (33) | 2/4 (50) | 0/2 (0) | ||||
| Moderate-severe deficit | 1/6 (17) | 1/4 (25) | 0/2 (0) | ||||
| Profound deficit | 3/6 (50) | 1/4 (25) | 2/2 (100) | ||||
| Sensory processing | DDf | DDf | DDf | ||||
| Dutch cohort (n = 13) | |||||||
| Tactile sensitivity | 4/6 (67) | 1/6 (17) | 2/4 (50) | 1/4 (25) | 2/2 (100) | 0/2 (0) | |
| Taste/smell sensitivity | 0/6 (0) | 2/6 (33) | 0/4 (0) | 2/4 (50) | 0/2 (0) | 0/2 (0) | |
| Movement sensitivity | 4/6 (67) | 0/6 (0) | 4/4 (100) | 0/4 (0) | 0/2 (0) | 0/2 (0) | |
| Under responsive/seeks sensation | 2/6 (33) | 1/6 (17) | 1/4 (25) | 1/4 (25) | 1/2 (50) | 0/2 (0) | |
| Auditory filtering | 0/6 (0) | 2/6 (33) | 0/4 (0) | 2/4 (50) | 0/2 (0) | 0/2 (0) | |
| Low energy/weak | 6/6 (100) | 0/6 (0) | 4/4 (100) | 0/4 (0) | 2/2 (100) | 0/2 (0) | |
| Visual/auditory sensitivity | 1/6 (33) | 1/6 (17) | 0/4 (0) | 1/4 (25) | 1/2 (50) | 0/2 (0) | |
| Behavioral questionnaires | |||||||
| Stereotypic movements | 20/31 (65) | 12/22 (55) | 8/9 (89) | 41/59 (69) | |||
| GERD behavior | 23/31 (74) | 16/22 (73) | 7/9 (78) | ||||
| Self-injurious behavior | 11/31 (35) | 8/22 (36) | 3/9 (33) | 47/61 (77) |
- Percentages are shown in brackets.
- Blank cells indicate that information was unavailable or uncertain.
- a Classification based on DC-LD, WHO and DSM-5.
- b Based on validated testing by behavioral specialist.
- c Physician reported data, no validated testing data available.
- d Number of individuals (of total individuals of whom are data available) who has acquired this milestone during given period of age at the time of present study.
- e Number of individuals (of total individuals of whom are data available) aged ≥ 5 years who has not acquired this skill at time of present study.
- f DD, Definite Difference; PD, Probable Difference; some individuals could not be assessed on Taste/Smell sensitivity and/or Movement sensitivity due to PEG tube and not able to move independently.
| SMC1A | NIPBL | |||
|---|---|---|---|---|
| All n = 51 | Missense variants n = 36 | Other variants n = 15 | All variants n = 67 | |
| Prenatal growth | ||||
| >2500 g | 26/41 (63) | 17/28 (61) | 9/13 (69) | 15/63 (24) |
| 1500–2500 g | 15/41 (37) | 11/28 (39) | 4/13 (31) | 37/63 (59) |
| <1500 g | 0/41 (0) | 0/28 (0) | 0/13 (0) | 11/63 (17) |
| Postnatal growthb | ||||
| >P75 | 27/38 (71) | 19/27 (70) | 8/11 (73) | 11/66 (17) |
| P25–P75 | 11/38 (29) | 8/27 (30) | 3/11 (27) | 41/66 (62) |
| <P25 | 0/38 (0) | 0/27 (0) | 0/11 (0) | 14/66 (21) |
| Head growth | ||||
| >−2SD | 15/37 (40) | 10/27 (37) | 5/10 (50) | 6/66 (9) |
| −2SD to −4SD | 17/37 (46) | 12/27 (44) | 5/10 (50) | 22/66 (33) |
| <−4SD | 5/37 (14) | 5/27 (19) | 0/10 (0) | 38/66 (58) |
| Limb malformationc | ||||
| No | 0/49 (0) | 0/35 (0) | 0/14 (0) | 50/67 (75) |
| Partial | 0/49 (0) | 0/35 (0) | 0/14 (0) | 4/67 (6) |
| Severe | 0/49 (0) | 0/35 (0) | 0/14 (0) | 13/67 (19) |
| Faced | ||||
| Possible CdLS | 18/51 (35) | 9/36 (25) | 9/15 (60) | 0/67 (0) |
| Mild | 24/51 (47) | 18/36 (50) | 6/15 (40) | 10/67 (15) |
| Classical | 9/51 (18) | 9/36 (25) | 0/15 (0) | 57/67 (85) |
| Intellectual disabilitye, f | ||||
| Normal-borderline | 3/32 (9) | 2/20 (10) | 1/12 (8) | 0/66 (0) |
| Mild-moderate | 16/32 (50) | 10/20 (50) | 6/12 (50) | 22/66 (33) |
| Severe-profound | 13/32 (41) | 8/20 (40) | 5/12 (42) | 44/66 (67) |
| Total severity scoreg | ||||
| Mean (range) | 9.4 (6–13) | 9.7 (6–13) | 9 (8–10) | 13.5 (8–18) |
- a Between brackets percentages for the characteristic within each (sub)group.
- b CdLS standard growth curves were used for postnatal height.
- c No = no reduction defect; partial = partial reduction defects (absence 1/2 fingers); severe = severe reduction defects (absence 3 or more fingers or complicated oligo-/polydactyly).
- d Possible CdLS; mild = mild type; classical = classical type.
- e Classification based on DC-LD, WHO and DSM-5.
- f Physician reported data, no validated testing data available.
- g Total severity score = Σ(prenatal growth + postnatal growth + head growth + limb malformation + face + intellectual/adaptive functioning) (based on Bhuiyan et al., 2006).
| Patient | SMC1A NL001 | SMC1A NL002 | SMC1A NL007 | SMC1A NL011 | SMC1A NL008 | Wenger et al. (2016) USA012 | Lebrun et al. (2015) | Goldstein et al. (2015) Patient A | Goldstein et al. (2015) Patient B | Summary |
|---|---|---|---|---|---|---|---|---|---|---|
| Gender | F | F | F | F | F | F | F | F | F | 0M/9F |
| Genotype | ||||||||||
| Exon | 1 | 2 | 5 | 15 | 16 | 10 | 11 | 18 | 24 | |
| Nucleotide change | 31A > T | 157dup | 694G > T | 2364del | 2421-?_2562 + ?del | 1636_1638delATT | 1911 + 1G>T | 2853_2856delTCAG | 3549_3552dupGGCC | |
| Amino acid change | Asn11Tyr | Thr53AsnfsX34 | Glu232* | Asn788Lysfs*10 | Leu808Argfs*6 | 546del | Thr638Valfs*48 | Ser951Argfs*12 | Ile1185Glyfs*23 | |
| Type | Missense | Frameshift | Nonsense | Frameshift | Frameshift | In-frame | Frameshift | Frameshift | Frameshift | |
| Growth | ||||||||||
| Prenatal growth | >2500 g | >2500 g | >2500 g | >2500 g | 1500–2500 g | >2500 g | NA | >2500 g | > 2500g | 1/8:1500–2500 g |
| 7/8: >2500 g | ||||||||||
| Postnatal growth# | >P75 | >P75 | P25–P75 | >P75 | >P75 | P25–P75 | <P25 | <P25 | P25–P75 | 2/9: <P25 |
| 3/9: P25–P75 | ||||||||||
| 4/9: >P75 | ||||||||||
| Head growth | >−2SD | >−2SD | −2/−4SD | −2/−4SD | >−2SD | −2/−4SD | −2/−4SD | >−2SD | >2SD | 5/9: >−2SD |
| 4/9: −2/−4SD | ||||||||||
| CdLS face | Mild | Possible | Possible | Possible | Possible | Possible | Possible | Possible | Absent | |
| Arched eyebrows | − | − | − | − | − | − | − | − | − | 0/9 |
| Synophrys | + | + | − | − | − | − | + | − | − | 3/9 |
| Long eyelashes | + | + | + | − | − | + | + | + | − | 6/9 |
| Long philtrum | − | − | + | − | − | NA** | − | + | − | 2/8 |
| Thin upper vermillion | − | − | − | − | − | + | + | − | − | 2/9 |
| Downturned corners mouth | + | + | + | + | − | + | − | + | − | 6/9 |
| Epilepsy | ||||||||||
| Generalized epilepsy | + | + | + | + | + | + | + | + | + | 9/9 |
| Onset | 2.5 m | 5 m | 4 m | 9 m | 2 m | 2 m | 1 m | 4 m | 17 m | 8/9 < 1yr |
| Refractory | + | + | + | + | + | + | + | + | − | 8/9 |
| Rett-like symptoms | ||||||||||
| Regression | NA@ | + | + | NA@ | + | NA@ | NA@ | + | − | 4/5 |
| Intellectual disability | Profound | Profound | Profound | Severe | Profound | Severe-profound | Severe-profound | Profound | NA | 8/8 Severe-profound |
| Stereotypic hand movements | + | + | + | + | + | − | + | + | + | 8/9 |
| Hand wringing | + | + | + | + | − | − | + | NA | NA | 5/7 |
| Disturbed respiratory control | − | − | − | NA | + | NA | NA | NA | NA | 1/4 |
- +, present; −, absent; NA, not (reliably) available; m, month(s); yr, year(s).
- # CdLS standard growth curves.
- ** could not be reliably scored due to microform cleft lip.
- @ could not reliably be scored due to early onset of seizures.
The congenital cardiac malformations observed in individuals with SMC1A mutations consisted of pulmonic stenosis (n = 3), atrial septal defects (n = 3), persistent ductus arteriosus (n = 2), ventricular septal defect (n = 1), dextrocardia (n = 1), aortic coarctation (n = 1), pulmonary valve dysplasia (n = 1), and left ventricular noncompaction with apical hypertrophy (n = 1). Cryptorchidism was scored as a minor anomaly and was present in four of the 15 males (27%) with SMC1A variants; 31/34 males (91%) with NIPBL variants had cryptorchidism. Early pubic hair development was reported in four females with a pathological SMC1A variant.
3.2 Milestones
While tabulating the milestones we left out SMC1A positive children below 5 years of age who were still too young to score with certainty whether they would or would not acquire the milestone before the age of 5 years. If a child ≥5 year old had not reached a milestone we indicated this.
3.3 Genotypes
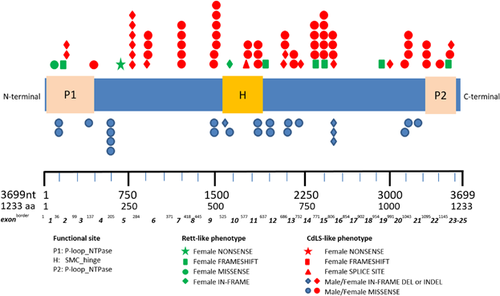
Of the present series half (26/51) of patients have been published before. The nature and site of variants in the present series does not differ from those reported in literature (Table 5; Figure 3).
| Index (reference) | Targeted analysis | Exon | Nucleotide change | Amino acid change | Coding effect | |
|---|---|---|---|---|---|---|
| Literature | ||||||
| 1 | BAB4135 Yuan, 2015b | – | 2 | c.121C>T | p.Leu41Phe | Missense |
| 2 | BAB4136 Yuan, 2015b | – | 2 | c.121C>T | p.Leu41Phe | Missense |
| 3 | Pt 3P Deardorff, 2007/Yuan, 2015 | + | 2 | c.173_187del | p.Val58_Arg62del | In-frame |
| 4 | Pt 2 Gervasini, 2013/Parenti, 2014 | + | 2 | c.173_187del | p.Val58_Arg62del | In-frame |
| 5 | Pt 4P Deardorff, 2007/Yuan, 2015 | + | 3 | c.397T>G | p.Phe133Val | Missense |
| 6 | Liu, 2009 | + | 3 | c.421G>A | p.Glu141Lys | Missense |
| 7 | Pt 2 Borck, 2007 | + | 4 | c.587G>A | p.Arg196His | Missense |
| 8 | Pt 5P Deardorff, 2007 | + | 4 | c.587G>A | p.Arg196His | Missense |
| 9 | Pie, 2010 | + | 4 | c.587G>A | p.Arg196His | Missense |
| 10 | Pie, 2010 | + | 5 | c.802_804del | p.Lys268del | In-frame |
| 11 | Liu, 2009 | + | 5 | c.802_804del | p.Lys268del | In-frame |
| 12 | Liu,2009 | + | 5 | c.802_804del | p.Lys268del | In-frane |
| 13 | BAB3623 Yuan, 2015 | – | 5 | c.802_804del | p.Lys268del | In-frame |
| 14 | Liu, 2009 | + | 5 | c.916_918del | p.Ser306del | In-frame |
| 15 | Pt 3 Gervasini, 2013 | + | 7 | c.1192C>G | p.Arg398Gly | Missense |
| 16 | Liu, 2009 | + | 7 | c.1193G>A | p.Arg398Gln | Missense |
| 17 | Liu, 2009 | + | 7 | c.1193G>A | p.Arg398Gln | Missense |
| 18 | Liu, 2009 | + | 7 | c.1193G>A | p.Arg398Gln | Missense |
| 19 | Pt II3 Musio, 2006 | + | 9 | c.1478A>C | p.Glu493Ala | Missense |
| 20 | Liu, 2009 | + | 9 | c.1478A>C | p.Glu493Ala | Missense |
| 21 | Liu, 2009 | + | 9 | c.1478A>C | p.Glu493Ala | Missense |
| 22 | Pt 6P Deardorff, 2007 | + | 9 | c.1486C>T | p.Arg496Cys | Missense |
| 23 | Pt 7P Deardorff, 2007b | + | 9 | c.1487G>A | p.Arg496His | Missense |
| 24 | Pt 7S Deardorff, 2007b | + | 9 | c.1487G>A | p.Arg496His | Missense |
| 25 | Pt 8P Deardorff, 2007b | + | 9 | c.1487G>A | p.Arg496His | Missense |
| 26 | Pt 8S Deardorff, 2007b | + | 9 | c.1487G>A | p.Arg496His | Missense |
| 27 | Pt 9P Deardorff, 2007 | + | 9 | c.1487G>A | p.Arg496His | Missense |
| 28 | Ansari, 2014 | – | 10 | c.1585_1587del | p.Lys529del | In-frame |
| 29 | Wenger, 2016 | – | 10 | c.1636_1638delATT | p.546del | In-frame |
| 30 | Hansen, 2013 | – | 10 | c.1731G>A | p.Glu577Glu | Splice defect |
| 31 | Ansari, 2014 | – | 11 | c.1757G>A | p.Arg586Gln | Missense |
| 32 | Lebrun, 2015 | – | 11 | c.1911 + 1G>T | p.Thr638Valfs*48 | Frameshift |
| 33 | Pt 17 Tzschach, 2015 | – | 12 | c.1937T>C | p.Phe646Ser | Missense |
| 34 | Pt 1 Gervasini, 2013 | + | 12 | c.1951G>A | p.Val651Met | Missense |
| 35 | Liu, 2009 | + | 12 | c.2046_2048delAGA | p.Glu683del | In-frame |
| 36 | Liu, 2009 | + | 12 | c.2077C>G | p.Arg693Gly | Missense |
| 37 | Pt 4 Gervasini, 2013/Parenti, 2014 | + | 13 | c.2078G>A | p.Arg693Gln | Missense |
| 38 | Pt 10P Deardorff, 2007 | + | 13 | c.2131C>T | p.Arg711Trp | Missense |
| 39 | Liu, 2009 | + | 13 | c.2131C>T | p.Arg711Trp | Missense |
| 40 | Pie, 2010 | + | 13 | c.2132G>A | p.Arg711Gln | Missense |
| 41 | Hoppman-Chaney, 2011 | + | 13-16 | c.2184_2563-268del | p.Leu729_Lys854delinsAspGluIle | In-frame |
| 42 | Liu, 2009 | + | 14 | c.2342G>T | p.Cys781Phe | Missense |
| 43 | Limongelli, 2010 | + | 15 | c.2351T>C | p.Ile784Thr | Missense |
| 44 | Pt 5 Gervasini, 2013/Parenti, 2014 | + | 15 | c.2351T>C | p.Ile784Thr | Missense |
| 45 | Pt 3 Fieremans, 2016 | – | 15 | c.2351T>C | p.Ile784Thr | Missense |
| 46 | Pt 26 De Ligt, 2012/ Pt 13 Gillissen, 2014/ Pt1 Jansen, 2016 | – | 15 | c.2364del | p.Asn788Lysfs*10 | Frameshift |
| 47 | Ansari, 2014 | – | 15 | c.2368C>T | p.Arg790Trp | Missense |
| 48 | Pt 11P Deardorff, 2007 | + | 15 | c.2369G>A | p.Arg790Gln | Missense |
| 49 | Ansari, 2014 | – | 15 | c.2369G>A | p.Arg790Gln | Missense |
| 50 | Pt 6 Gervasini, 2013 | + | 15 | c.2369G>A | p.Arg790Gln | Missense |
| 51 | Pt 98 De Ligt, 2012/ Pt 48 Gillissen, 2014 / Pt2 Jansen, 2016 | – | 16-17 | c.2421_2652del | p.Leu808Argfs*21 | Frameshift |
| 52 | Liu, 2009 | + | 16 | c.2446C>G | p.Arg816Gly | Missense |
| 53 | Mannini, 2010 | + | 16 | c.2467T>C | p.Phe823Leu | Missense |
| 54 | Pt II4 Musio, 2006/Parenti, 2014b | + | 16 | c.2493_2495del | p.Asp831_Gln832delinsGlu | In-frame |
| 55 | Pt III2 Musio, 2006/Parenti, 2014b | + | 16 | c.2493_2495del | p.Asp831_Gln832delinsGlu | In-frame |
| 56 | Pt III3 Musio, 2006/Parenti, 2014b | + | 16 | c.2493_2495del | p.Asp831_Gln832delinsGlu | In-frame |
| 57 | Pt III4 Musio, 2006 /Parenti, 2014b | + | 16 | c.2493_2495del | p.Asp831_Gln832delinsGlu | In-frame |
| 58 | Pt A Goldstein, 2015 | – | 18 | c.2853_2856delTCAG | p.Ser951Argfs*12 | Frameshift |
| 59 | BAB5452 Yuan, 2015 | – | 19 | c.2974_2A>G | p.Asp992_Gln994del | In-frame |
| 60 | Liu, 2009 | + | 20 | c.3146G>A | p.Arg1049Gln | Missense |
| 61 | Jang, 2015b | – | 21 | c.3178G>A | p.Glu1060Lys | Missense |
| 62 | Jang, 2015b | – | 21 | c.3178G>A | p.Glu1060Lys | Missense |
| 63 | Jang, 2015b | – | 21 | c.3178G>A | p.Glu1060Lys | Missense |
| 64 | Jang 2015b | – | 21 | c.3178G>A | p.Glu1060Lys | Missense |
| 65 | Pt 1 Borck, 2007 | + | 21 | c.3254A>G | p.Tyr1085Cys | Missense |
| 66 | Pt 12P Deardorff, 2007 | + | 22 | c.3364T>C | p.Phe1122Leu | Missense |
| 67 | Liu, 2009 | + | 22 | c.3367C>T | p.Arg1123Trp | Missense |
| 68 | Pt 7 Gervasini, 2013/Parenti, 2014 | + | 23 | c.3497A>C | p.Asn1166Thr | Missense |
| 69 | Pat B Goldstein, 2015 | – | 24 | c.3549_3552dupGGCC | p.Ile1185Glyfs*23 | Frameshift |
| 70 | Pt 8 Gervasini, 2013/Parenti, 2014 | + | 24 | c.3565C>T | p.Leu1189Phe | Missense |
| 71 | Ansari, 2014 | – | 24 | c.3574_3576del | p.Glu1192del | In-frame |
| 72 | Baquero, 2014)/Pie, 2016 | + | 1-25 | Dup Xp11.22 region ∼1.1Mb | ||
| Present series | ||||||
| 1 | SMC1ANL001c | + | 1 | c.31A>T | p.Asn11Tyr | Missense |
| 2 | SMC1ANL002c | + | 2 | c.157dup | p.Thr53AsnfsX34 | Frameshift |
| 3 | SMC1AUSA004 (Deardorff, 2007) | 2 | c.173_187del | p.Val58_Arg62del | In-frame | |
| 4 | SMC1AUSA008 (Deardorff, 2007) | 3 | c.397T>G | p.Phe133Val | Missense | |
| 5 | SMC1ASPA001 (Deardorff, 2007) | + | 4 | c.587G>A | p.Arg196His | Missense |
| 6 | SMC1AGER003 | + | 4 | c.587G>A | p.Arg196His | Missense |
| 7 | SMC1AFR003 (Borck, 2007) | + | 4 | c.587G>A | p.Arg196His | Missense |
| 8 | SMC1ANL007c | + | 5 | c.694G>T | p.Glu232* | Nonsense |
| 9 | SMC1ADEN001 | + | 5 | c.802_804del | p.Lys268del | In-frame |
| 10 | SMC1ASPA002 (Pie, 2010) | + | 5 | c.802_804del | p.Lys268del | In-frame |
| 11 | SMC1AUK008 | + | 5 | c.802_804del | p.Lys268del | In-frame |
| 12 | SMC1AUSA002 (Liu, 2009) | 5 | c.802_804del | p.Lys268del | In-frame | |
| 13 | SMC1AFR005 | + | 6 | c.919C>A | p.His307Asn | Missense |
| 14 | SMC1ADEN002 | + | 6 | c.920A>T | p.His307Leu | Missense |
| 15 | SMC1AGER004/SMC1AARG001 | ? | 7 | c.1193G>A | p.Arg398Gln | Missense |
| 16 | SMC1AGER001/SMC1AAUSTR001 | + | 9 | c.1475A>G | p.Gln492Arg | Missense |
| 17 | SMC1ADEN003 b/SMC1AUSA007 b (Deardorff, 2007) | + | 9 | c.1487G>A | p.Arg496His | Missense |
| 18 | SMC1ADEN004 b/SMC1AUSA006 b (Deardorff, 2007) | + | 9 | c.1487G>A | p.Arg496His | Missense |
| 19 | SMC1AUSA001 (Deardorff, 2007) | 9 | c.1487G>A | p.Arg496His | Missense | |
| 20 | SMC1AUK002 (Ansari, 2014) | + | 10 | c.1585_1587 del | p.Lys529del | In-frame |
| 21 | SMC1AUK006 | + | 10 | c.1607A>T | p.Lys536Met | Missense |
| 22 | SMC1AUSA012 (Wenger, 2016) | – | 10 | c.1636_1638delATT | p.546del | In-frame |
| 23 | SMC1AUSA010 | 11 | c.1756C>T | p.Arg586Trp | Missense | |
| 24 | SMC1AUK004 (Ansari, 2014) | + | 11 | c.1757C>T | p.Arg586Gln | Missense |
| 25 | SMC1ANL009b | + | 11 | c.1847C>A | p.Ala616Asp | Missense |
| 26 | SMC1ANL010b | + | 11 | c.1847C>A | p.Ala616Asp | Missense |
| 27 | SMC1ANL006 | – | 11 | c.1904G>A | p.Arg635His | Missense |
| 28 | SMC1ANL014b | – | 11 | c.1904G>A | p.Arg635His | Missense |
| 29 | SMC1ANL015b | – | 11 | c.1904G>A | p.Arg635His | Missense |
| 30 | SMC1AGER002/SMC1ASWI001 | + | 13 | c.2078G>A | p.Arg693Gln | Missense |
| 31 | SMC1AFR004 | + | 13 | c.2090_2092dup | p.Glu697_Leu698delinsVal | In-frame |
| 32 | SMC1ANL005 | + | 13 | c.2095C>T | p.Arg699Cys | Missense |
| 33 | SMC1AUSA005 (Deardorff, 2007) | 13 | c.2131C>T | p.Arg711Trp | Missense | |
| 34 | SMC1ASPA003 (Pie, 2010) | + | 13 | c.2132G>A | p.Arg711Gln | Missense |
| 35 | SMC1AITA003 (Gervasini, 2013) | + | 15 | c.2351T>C | p.Ile784Thr | Missense |
| 36 | SMC1ANL011 (Jansen, 2016) | – | 15 | c.2364del | p.Asn788Lysfs*10 | Frameshift |
| 37 | SMC1AUK001 (Ansari, 2014) | + | 15 | c.2368C>T | p.Arg790Trp | Missense |
| 38 | SMC1ASPA004 (Deardorff, 2007) | + | 15 | c.2369G>A | p.Arg790Gln | Missense |
| 39 | SMC1AUK007/SMC1AIND001 (Ansari, 2014) | + | 15 | c.2369G>A | p.Arg790Gln | Missense |
| 40 | SMC1AUK005/SMC1ATUR001 | + | 15 | c.2369G>A | p.Arg790Gln | Missense |
| 41 | SMC1ANL008 (Jansen, 2016) | – | 16 | c.2421-?_2562+?del | p.Leu808Argfs*6 | Frameshift |
| 42 | SMC1AUSA011 (Liu, 2009) | 16 | c.2446C>G | p.Arg816Gly | Missense | |
| 43 | SMC1AFR001 | + | 16 | c.2455A>C | p.Ile819Leu | Missense |
| 44 | SMC1AITA001b (Musio, 2006) | + | 16 | c.2493_2495del | p.Asp831_Gln832delinsGlu | In-frame |
| 45 | SMC1AITA002b | + | 16 | c.2493_2495del | p.Asp831_Gln832delinsGlu | In-frame |
| 46 | SMC1ANL004 | + | 21 | c.3145C>G | p.Arg1049Gly | Missense |
| 47 | SMC1AFR0021 (Borck, 2007) | + | 21 | c.3254A>G | p.Tyr1085Cys | Missense |
| 48 | SMC1AUSA003 (Deardorff, 2007) | 22 | c.3364T>C | p.Phe1122Leu | Missense | |
| 49 | SMC1ANL003c | + | 22 | c.3367C>T | p.Arg1123Trp | Missense |
| 50 | SMC1AITA004 (Gervasini, 2013) | + | 23 | c.3497A>C | p.Asn1166Thr | Missense |
| 51 | SMC1AUK003 (Ansari, 2014) | + | 24 | c.3574_3576del | p.Glu1192del | In-frame |
- Blank cell indicates that information was unavailable or uncertain.
- a Annotation according to reference sequence NM_006306.3.
- b Familial cases.
- c Panel analysis (epilepsy, Rett syndrome); clinically the patients were not suspected as having CdLS, other diagnoses were thought to be more likely.
3.4 Reasons for molecular analysis
In the Dutch cohort, 5/15 (38%) of patients were clinically suspected of CdLS prior to molecular testing. For five patients CdLS was included in the differential diagnosis, but other diagnoses were thought to be more likely. For the remaining three patients CdLS was not clinically suspected at all. All patients coming from other countries were clinically suspected to have CdLS prior to molecular testing. The testing methods differed among patients depending on local laboratory protocols, and included Sanger sequencing, panel analysis aimed at genes associated with CdLS, and panel analysis aimed at genes associated with intellectual disability/epilepsy.
4 DISCUSSION
SMC1A is known as a gene that can cause a cohesinopathy if mutated (Musio et al., 2006). The entities tagged as cohesinopathies have been considered overlapping entities (Liu & Krantz, 2008). They share several physical and behavioral features, such as limited growth, several of the facial features, limb malformations, and intellectual disability. The cohesin complex and its regulators mediate sister-chromatid cohesion in dividing cells and are important for controlling gene expression (Remeseiro, Cuadrado, & Losada, 2013). Sharing of major features of the cohesinopathies supports the hypothesis that a disturbed cohesin function contributes to these characteristics (Yuan et al., 2015). There are also differences in the phenotypes caused by SMC1A and NIPBL pathological variants. Such differences support the argument that the phenotype is not only a result of the disturbed cohesin function, but also a result of other functions (moonlighting) of the cohesin genes (Jeffery, 2014). One major difference in phenotype between the SMC1A and NIPBL group described here is the higher prevalence and more severe form of self-injurious behavior in the latter. The absence of this behavioral trait in patients with SMC1A variants with a Rett-like phenotype, and also in other cohesinopathies, such as individuals with CdLS due to variants in other genes and in individuals with Roberts syndrome (Vega et al., 2005), suggests a moonlighting hypothesis for NIPBL. Indeed knock-out mouse models for Nipbl have shown that Nipbl affects transcription and global dysregulation of gene expression, and consequently does have functions different from the cohesin function and have shown evidence for different polypeptide chain functions of NIPBL products and for expression changes in genes with roles in neuronal functions that underlie the behavioral and neurological abnormalities observed (Kawauchi et al., 2009, 2016).
Patients with cohesinopathies share several physical signs and symptoms that have been implicated as cause of SIB (Luzzani, Macchini, Valade, Milani, & Selicorni, 2003), and this argues against the self-injurious behavior being secondary to these physical conditions. Therefore, further studies into cohesinopathies and their associated genes, should not only be aimed at the cohesin and related functions, but should also take into account other potential functions of these genes.
The higher incidence of SIB in the NIPBL group could be due to the cognitive level, since cognitive functioning is overall more affected in the NIPBL group than in the SMC1A group. However, SIB seems to be absent in the Rett-like group and yet cognitive functioning appears even lower. Further developmental testing may indicate other cognitive and behavioral differences that may contribute to this. An association (if any) between the results of cognitive and developmental assessments and SIB, and results of the behavioral studies should be described in much detail and will therefore be published elsewhere ().
SMC1A variants are known to be associated with a CdLS phenotype. In comparing CdLS characteristics in the present study, the SMC1A group demonstrates a less disturbed growth compared to the NIPBL group. Prenatal growth parameters are below 2 SD in one-third of the SMC1A group, irrespective of the mutation type. In the NIPBL group prenatal growth parameters are below 2 SD in at least two-thirds of the group. Postnatal height and occipitofrontal circumference are decreased in two-thirds of the SMC1A group, which is less marked compared to the NIPBL group. However, weight is much more disturbed in the NIPBL group, possibly due to the much more frequent, more severe and more protracted feeding problems in this group.
All facial features that characterize CdLS can be present in individuals with SMC1A variants, but in a lower frequency compared to the NIPBL group. There are some exceptions: individuals with a missense SMC1A variant have the same frequency of periocular features as individuals in the NIPBL group, and also the prevalence of the thin upper vermillion is similar between the two groups. CdLS features that are more prevalent in the NIPBL group such as a small lower jaw and low-set and malformed ears occur more frequently in the group with a missense SMC1A mutation than in the group with other mutation types. However, the number of individuals in the latter group is small and results should be evaluated with care.
Limb reduction defects that are typical for CdLS and prevalent in 25% of the NIPBL group, are absent in the SMC1A group. Clinodactyly of the fifth finger occurs less frequently (X2 p = 0.038) than in the NIPBL group, and small hands and a proximally placed thumb are also less frequent (statistically not significant). Feeding problems are more frequent in the NIPBL group (X2 p = 0.0001), while gastroesophageal reflux disease and constipation are equally common in both groups. Seizures, however, are more frequent in the SMC1A group (X2 p = 0.0005), and this is more marked in the group with non-missense SMC1A variants (statistically not significant).
A comparison of cognition and behavior is hampered by the lack of data in a considerable number of individuals in the international SMC1A group and the NIPBL group. The numbers of the in person tested individuals in the Dutch cohort are small and should be used with care. All tested individuals in the Dutch cohort have problems with sensory processing.
In summary, individuals with SMC1A variants show a phenotype that overlaps with CdLS. The frequencies of some signs and symptoms are lower than in individuals with NIPBL mutations. Major phenotypic distinctions are the absence of limb reduction defects and increased prevalence of seizures in the SMC1A group. Another main difference is self-injurious behavior which is much more frequent and more severe in the NIPBL group.
The Dutch SMC1A group likely covers all individuals with SMC1A variants currently known in the Netherlands. The group includes both patients who were clinically diagnosed with CdLS, and those in whom a variant was unexpectedly detected through exome sequencing. We recognize two groups in the Dutch cohort: individuals with a phenotype similar to CdLS, and a group with an epileptic encephalopathy. Individuals with an epileptic encephalopathy have been previously reported as well (de Ligt et al., 2012; Fieremans et al., 2016; Gilissen et al., 2014; Goldstein et al., 2015; Hansen et al., 2013; Jang et al., 2015; Jansen et al., 2016; Lebrun et al., 2015; Tzschach et al., 2015). In the Dutch cohort 5 of 13 (38%) individuals had an epileptic encephalopathy. In evaluating these female patients we were struck by the resemblance to females with progressed stages in Rett syndrome and their typical impaired ability to make contact and interact. All have severe or profound intellectual disabilities and four of the five Dutch females (five of the seven females of the total Rett-like group) showed hand movements such as “hand wringing” (Table 4). Regression has been reported in literature (Goldstein et al., 2015; Jansen et al., 2016) and is reported here in three of the five females (Table 4). In two other females epilepsy and developmental delay manifested at such young age that this may have masked any sign of regression. Other characteristics of the individuals with an epileptic encephalopathy were a lower birth weight and a lower postnatal height compared to the others in the SMC1A group. According to severity classification terminology (classical, mild, possible CdLS) their faces were assessed as possible CdLS, except in the youngest female who was assessed as mild CdLS. No face morphology was rated as classical CdLS. There is anecdotal evidence that individuals with SMC1A variants have a rounder face compared to individuals with NIPBL variants and this seems more marked in individuals with a Rett-like phenotype than in individuals with SMC1A variants in general (Figure 1).
We considered a cluster analysis of signs and symptoms to determine which set of phenotypical characteristics is more similar to each other in one sub-phenotype than in another, but the total numbers were too small to allow for meaningful results.
The exact phenotype of the subgroup of individuals with SMC1A variants with an epileptic encephalopathy and severe-profound intellectual disability has not emerged yet, but it is likely that more individuals will be recognized as exome sequencing is increasingly used worldwide. This may allow better insight whether the phenotypes are truly separate or rather ends of a spectrum. In the Netherlands, five of the thirteen patients known with SMC1A pathological variants have an epileptic encephalopathy phenotype (Table 4). Possibly this phenotype is much more common than anticipated. The mutations of individuals with the epileptic encephalopathy are spread all over the gene and a clear correlation does not appear (Figure 3). All mutations are nonsense or frameshift mutations except one missense mutation (SMC1ANL001; Table 4), located at the first part of exon 1, in which functional studies have indicated it to cause a loss of function as well (Dr. Erwan Watrin, personal communication, 2017). To date there is no known exon 8 SMC1A mutation.
SMC1A incompletely escapes X-inactivation (Gervasini et al., 2013; Goldstein et al., 2015; Limongelli et al., 2010; Liu, Feldman, et al., 2009; Mannini et al., 2010; Tzschach et al., 2015). Since, there is no altered level of SMC1A transcripts and mutant proteins maintain a residual function (Liu, Feldman, et al., 2009), and a dominant negative effect is considered the pathogenic mechanism in females with a SMC1A variant, the level of allelic preferential expression might be one of the factors contributing to the wide phenotypic variability observed in these patients (Parenti et al., 2014). In the present study there is a remarkably distorted ratio of males and females with a SMC1A variant for non-missense variants. The small number of males with non-missense variants had in frame deletions. This seems to indicate that other types of mutations are not tolerated in males, likely leading to early miscarriages, and explaining the distorted gender ratio. We evaluated spontaneous abortions reported by the families: 22/49 (45%) families reported no known miscarriages, 3/49 (6%) families experienced a single miscarriage, and one (2%) family (with mutation c.3145C>G; p.Arg1049Gly) had six miscarriages for which no cause could be found (no data on the other 24 families). Although, normal values for spontaneous miscarriages in the various populations are not available it seems likely the miscarriage rate in the families in total is not increased.
The present study has several limitations. First, the CdLS-like phenotype in the SMC1A group is very likely overestimated due to acquisition bias, as patients suspected to have CdLS were referred to CdLS specialists, whom we specifically invited to participate in the study. The specialists confirmed that all included individuals with SMC1A variants were suspected to have CdLS. We contacted the UK 100,000 genome project in order to obtain an estimate of the frequency of SMC1A mutations in a large group of individuals, but at present such a detailed question cannot be answered yet (Richard Scott, personal communication, 2016). Therefore, the phenotype presented here is mainly representative of the phenotype similar to CdLS and less of the epileptic encephalopathy “Rett-like” phenotype. Although, numbers are small, prevalences of these two subphenotype groups in the Netherlands indicate that the latter phenotype might occur even more frequently than the former.
Furthermore, cross sectional data collection using binary categories to describe features hampers the reporting of gradations and changes over time. Moreover, as the somatic questionnaire was extensive, we had to deal with missing data from several patients. These experiences underline the importance of using standardized, longitudinal databases (Baas et al., 2015). Performing research with a large group of collaborating physicians may have influenced phenotype evaluations, especially with respect to facial morphology. As differences between the presently in person examined patients and patients evaluated by a group of others were small, it seems unlikely that this has played a major role.
Data on cognitive and adaptive functioning and the measures used are often missing in the medical file, and if these are available, different developmental and behavioral assessment instruments are typically used. We strongly advocate direct and indirect assessments of cognitive and adaptive functioning and behavior of affected individuals, performed by behavioral scientists, and that these always form an integrated part of an interdisciplinary evaluation.
We conclude that SMC1A variants can result in different phenotypes: a phenotype that overlaps with mild manifestations of CdLS and one that overlaps with Rett syndrome. Likely the increasing use of exome and genome sequencing will lead more frequently to identification of SMC1A variants in individuals not clinically suspected of CdLS. Large series of individuals recognized in this way should facilitate cluster analyses that may allow either separating distinct SMC1A phenotypes or merging these into one spectrum. Such better insights will allow better genetic counseling, allow health care professionals to answer the primary question of parents what it means if a SMC1A variant is found in their child.
ACKNOWLEDGMENTS
We are exceptionally grateful to the patients with SMC1A variants and their families who participated in this study. We are very grateful to the Prinsenstichting for funding in part the work of SH, and to the Dutch and Polish CdLS Associations for cooperation in the development of detailed clinical data NIPBL positive patients. We sincerely thank Dr Alina Kuzniacka and Natalia Krawczyńska from Department of Biology and Genetics, Medical University in Gdańsk for molecular analysis. This work was supported by National Institutes of Health Grants UMO-2014/15/B/NZ5/03503. This work was supported by the Spanish Ministry of Health − ISCIII, Fondo de Investigación Sanitaria (FIS) [Ref: PI15/00707] and the Diputación General de Aragón [Grupo Consolidado B20], European Social Fund (“Construyendo Europa desde Aragón”) to FJRF and JPJ. This work was funded by the German Federal Ministry of Education and Research (BMBF, CHROMATIN-Net) to FJK and GG-K. We dedicate this manuscript to the excellent clinician and caregiver, a colleague and a friend, Ton van Essen, who died during the preparation of the manuscript.