1q21.3 deletion involving GATAD2B: An emerging recurrent microdeletion syndrome
Abstract
GATAD2B gene is involved in chromatin modification and transcription activity. Loss-of-function mutations of GATAD2B have recently been defined to cause a recognizable syndrome with intellectual disability (ID). Human TPM3 gene encoding thin filament protein is associated with myopathies. Both genes are located on chromosome 1q21.3. We herein report an infant with feeding difficulty, developmental delay, hypotonia, and dysmorphic features including small palpebral fissures, telecanthus, sparse hair and eyebrow, cup-shaped ears, and clinodactyly. Karyotype was normal. Single nucleotide polymorphism array revealed a 1.06 Mb deletion of chromosome 1q21.3, which was confirmed to be de novo. The deleted region encompassed 35 genes, including three known disease-associated genes, namely GATAD2B, TPM3, and HAX1. We further identify and summarize seven additional patients with 1q21.3 microdeletion from literature review and clinical databases (DECIPHER, ISCA/ClinGen). Genomic location analysis of all eight patients revealed different breakpoints and no segmental duplication, indicating that non-homologous end joining is a likely mechanism underlying this particular microdeletion. This data suggests that 1q21.3 microdeletion is a recurrent microdeletion syndrome with distinguishable phenotypes, and loss of function of GATAD2B is the major contributor of the characteristic facies and ID. Additionally, the deletion of TPM3 warrants a risk of concomitant muscle disease in our patient. © 2017 Wiley Periodicals, Inc.
INTRODUCTION
The human GATA zinc finger domain containing 2B (GATAD2B) gene encodes beta-subunit of the transcription repressor complex MeCP1-Mi2/nucleosome remodeling and deacetylase complex involved in chromatin modification and transcription activity. Recently, loss-of-function mutations of GATAD2B including three point mutations and one chromosomal microdeletion were defined to be a cause of severe intellectual disability (ID) [de Ligt et al., 2012; Hamdan et al., 2014] and recognizable syndrome characterized by childhood hypotonia and dysmorphic features including broad forehead, thin hair, narrow palpebral fissure, tubular shaped nose with broad tip, deeply set eyes, short philtrum, broad mouth, strabismus, and long fingers [Willemsen et al., 2013]. Human tropomyosin 3 (TPM3) gene encodes muscle thin filament protein. Mutations of the TPM3 gene are associated with three distinct autosomal dominant and recessive myopathies, namely nemaline myopathy, congenital fiber-type disproportion, and cap myopathy [Wattanasirichaigoon et al., 2002; Penisson-Besnier et al., 2007; Citirak et al., 2014]. Herein, we describe a patient of 1q21.3 microdeletion containing GATAD2B and TPM3 genes with expanded phenotypes. Furthermore, seven additional patients with 1q21.3 microdeletion were reviewed from literature, DECIPHER and ISCA/ClinGen databases. Phenotype and genomic location of this recurrent microdeletion syndrome were compared and genetic mechanism underlying this microdeletion was proposed.
CLINICAL REPORT
The patient was born via cesarean section, at 39-week gestation, with asymmetrical small for gestational age, birth weight of 2,130 g, length of 46 cm, and head circumference of 33 cm. Apgar scores were 7 and 9, at 1 and 5 min, respectively. The father and mother were 35-year-old with otherwise unremarkable family history. Chromosome analysis of amniotic fluid showed 46,XX, normal female karyotype. After discharged home on DOL3, the patient was noticed to have floppiness, feeding difficulty, and poor weight gain.
At 3 months of age, the patient was evaluated for developmental delay. Physical examination revealed body weight of 3,980 g (<3rd centile), length of 55 cm (10th centile), and minor dysmorphic features including high forehead, thin and light brown hair and eyebrows, telecanthus, narrow palpebral fissure, thin lips, low set and abnormal ear shape, widely spaced nipples, and bilateral clinodactyly of the 5th fingers (Fig. 1A). Her facial dysmorphism became more obvious including tented mouth appearance, telecanthus with epicanthal fold, and blunt nasal tip noted at 18 months (Fig. 1B). Neurological examination, at 22 months, revealed truncal hypotonia, and normal deep tendon reflex without limb weakness.
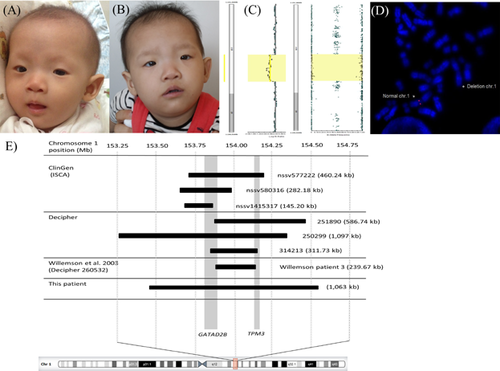
Ophthalmological examination revealed hyperopia and strabismus. An auditory brainstem response (ABR) and auditory steady state response (ASSR) showed mild hearing loss in the left ear. Cranial and renal ultrasounds were unremarkable.
In parallel, gastrostomy tube was placed at age 12 months, for feeding difficulty and gastroesophageal reflux, which led to improvement of weight gain and normalization of body weight (9.4 kg, 25th centile) at age 17 months. Her head size had been at 25th–50th centile since birth.
Developmentally, she could support her head at 6 months of age, vocalize and roll over at 9 months, babble and sit unsupported at 22 months. At age 30 months, she did not have meaningful words.
Rapid detection of 24 chromosomal abnormalities and nine microdeletion syndromes by BACs-on-Beads revealed disomy of the arm specific regions in all 22 autosomes with XX.
A single nucleotide polymorphism array (Illumina Infinium CytoSNP-850 K BeadChip) was performed and analyzed using BlueFuse Multi software v4.1, after obtaining a written informed consent, which revealed a 1.06 Mb deletion of chromosome 1q21.3 [Chr1:153,460,453–154,523,588, GRCh37] (Fig. 1C). The deleted fragment encompassed 35 genes, including S100A1, S100A2, S100A3, S100A4, S100A5, S100A7, S100A13, S100A14, S100A16, CHTOP, SNAPIN, ILF2, NPR1, INTS3, SLC27A3, GATAD2B, DENND4B, CRTC2, SLC39A1, CREB3L4, JTB, RAB13, RPS27, NUP210L, TPM3, C1orf43, C1orf189, UBAP2L, HAX1, AQP10, ATP8B2, IL6R, SHE, TDRD10, and UBE2Q1. Among these, only GATAD2B, TPM3, and HAX1 genes are known to be associated with human disorders. The 1q21.3 microdeletion was confirmed by metaphase fluorescence in situ hybridization (FISH) using BAC probe, RP11-216N14 (Fig. 1D). Parental karyotype and FISH analysis yielded normal findings.
We performed a review of literature and curated clinical copy number variation (CNV) databases (DECIPHER, ISCA/ClinGen) and identified one and six additional patients of 1q21.3 microdeletion involving GATAD2B, respectively. The phenotypes and overlapping genomic locations of all eight patients including our patient are summarized in Table I and Figure 1E [Firth et al., 2009; Willemsen et al., 2013; Rehm et al., 2015].
| Database/source | Variant ID | Location (GRCh37) | Deletion size (kb) | Phenotype | Inheritance | Numbers of gene involved | GATAD2B deletion | TPM3 deletion | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | The present patient | chr1:153460453-154523588 | 1,063 | DD, hypotonia, feeding difficulty, high forehead, thin and light hair/eyebrows, telecanthus, narrow palpebral fissure, thin lips, low set ear | de novo | 35 | Complete | Complete | |
| 2 | ClinGen (ISCA) | nssv 577222 | chr1:153732039-154192279 | 460 | Muscular hypotonia, microcephaly | NA | 14 | Complete | Complete |
| 3 | ClinGen (ISCA) | nssv 580316 | chr1:153671910-153954090 | 282 | DD and additional significant morphological phenotypesa | NA | 9 | Complete | No |
| 4 | ClinGen (ISCA) | nssv 1415317 | chr1:153724807-153870005 | 145 | DD and additional significant morphological phenotypesa | NA | 3 | Partial | No |
| 5 | Decipher | 251890 | chr1:153885226-154471969 | 586 | ID, hypotonia, macrocephaly, frontal bossing, micrognathia, hypertelorism, deeply set eye, overlapping fingers and long phalanx, strabismus, talipes equinovarus, abnormal palmar crease, slender build | de novo | 26 | Partial | Complete |
| 6 | Decipher | 250299 | chr1:153262829-154359839 | 1,097 | NA | NA | 50 | Complete | Complete |
| 7 | Decipher | 314213 | chr1:153835538-154147267 | 311 | NA | de novo | 14 | Partial | Partial |
| 8 | Willemsen et al. [2013] | Patient 3 | chr1:153893110-154132780 | 240 | DD, hypotonia, feeding difficulty, broad forehead, thin /blond hair, strabismus, hypermetropia, hypertelorism, broad/flat nasal bridge, full square tip of the nose | de novo | 10 | Partial | Partial |
- DD, developmental delay; ID, intellectual disability; NA, no data available.
- a No detailed morphological phenotype available.
DISCUSSION
We describe expanded phenotypes including, hearing loss, widely spaced nipples, and tented mouth appearance in GATAD2B-associated 1q21.3 microdeletion syndrome. We also note that tubular nose, broad nasal tip, and long-slender fingers are inconsistent findings in GATAD2B-associated ID syndrome since they were not seen in this patient [Willemsen et al., 2013]. However, given the younger age of this patient, further follow up of clinical phenotype is required.
The complete deletion of GATAD2B in the present patient supports previous observation that loss of function of this gene is the major contributor of the characteristic facial phenotype and ID. Study in fruit fly model showed that GATAD2B haploinsufficiency led to impaired habituation, a learning and memory phenotype in fruit fly model of ID, and disrupted synaptic development [Willemsen et al., 2013]. It should be mentioned that a de novo GATAD2B splicing mutation was identified in another child with ID, unfortunately there was no data on facial phenotype for comparison [Hamdan et al., 2014].
Based on the present patient and seven additional patients from literature/clinical databases, we propose that the 1q21.3 microdeletion involving GATAD2B is an emerging recurrent microdeletion syndrome [Firth et al., 2009; Willemsen et al., 2013; Rehm et al., 2015]. The sizes of 1q21.3 microdeletion involving GATAD2B were in the range of 145–1,093 kb [Firth et al., 2009; Willemsen et al., 2013; Rehm et al., 2015]. Among these, four had a complete GATAD2B deletion, and the remainders had a partial deletion of GATAD2B. The mechanism of 1q21.3 microdeletion is most likely due to non-homologous end joining (NHEJ) as there is no segmental duplication in this region of the genome. Additionally, the fact that all overlapping deletions seen in these patients do not share the same breakpoints is in line with NHEJ (Fig. 1E). We have also investigated whether there are recurrent reciprocal duplications in this region, and found that most duplications involving GATAD2B in the clinical CNV databases are large in size, ranging from 2 to 100 Mb, and not specific to 1q21.3 (data not shown). Hence, recurrent 1q21.3 microdeletions seem to be the main alteration in GATAD2B-related disorder.
Human TPM3 gene is distally located to GATAD2B on chromosome 1q21.3, with a distance of 230 kb in between. TPM3 mutations can result in nemaline myopathy, congenital fiber-type disproportion, and cap myopathy, all of these disorders show no cognitive involvement [Wattanasirichaigoon et al., 2002; Penisson-Besnier et al., 2007; Citirak et al., 2014]. Most, but not all, heterozygous TPM3 missense mutations lead to childhood onset muscle weakness, whereas biallelic loss-of-function mutation results in an infantile-onset muscle disease [Citirak et al., 2014; Marttila et al., 2014]. Six out of the eight 1q21.3 microdeletion patients possess partial or complete TPM3 deletion (Fig. 1E). Of these six patients, four had hypotonia, whereas data was unavailable in the remainders. For the two patients without TPM3 deletion, there was no detailed phenotype other than developmental delay (Table I). Based on the existing data, it is too premature to draw a conclusion on the phenotypic effect of TPM3 haploinsufficiency associated with 1q21.3 microdeletion. Although heterozygous TPM3 deletion is unlikely to result in infantile myopathy, it warrants a close and further follow up for possible myopathy later in life. Therefore, a larger number of 1q21.3 microdeletion patients with or without TPM3 deletion and long-term follow up till adulthood is required to shed light on the question. As for another disease-causing gene deleted in this patient, HAX1, the only disorder associated with HAX1 mutation is autosomal recessive congenital neutropenia, of which is not a concerned phenotype in our patient [Klein et al., 2007].
In conclusion, microdeletion of 1q21.3 is an emerging recurrent microdeletion syndrome. The possibility of NHEJ as an underlying mechanism leading to the microdeletion requires further investigation. Loss of function of GATAD2B is the major contributor of the shared facial features, hypotonia, developmental delay, and ID. Delineating the breakpoints and genes contained in the deletion fragment are essential for predicting the phenotypic variability. In our patient, the deletion of TPM3 warrants a risk of concomitant muscle disorder.
ACKNOWLEDGMENTS
We would like to thank the patient's family for their participation in this project, and Dr. Chintana Tocharoentanaphol for laboratory assistance. The work was supported by grants from Development Potentials of Thai People Project, Faculty of Medicine Ramathibodi Hospital (to NJ), and Mahidol University (to DW). DW, NJ, and TT are recipients of the Research Career Development Awards from the Faculty of Medicine at Ramathibodi Hospital. The dataset(s) used for the analyses described in this manuscript were obtained from the database of Genotype and Phenotype (dbGaP) found at http://www.ncbi.nlm.nih.gov/gap (dbGaP accession phs000205.v6.p2). Samples and associated phenotype data for the International Standards for Cytogenomic Arrays were provided by ISCA member laboratories.