A novel AMPD2 mutation outside the AMP deaminase domain causes pontocerebellar hypoplasia type 9
TO THE EDITOR
Pontocerebellar hypoplasia type 9 (PCH9, OMIM#615809) is a rare, fully penetrant, autosomal recessive neurodegenerative disorder with prenatal onset caused by variants affecting the function of the protein encoded by the gene adenosine monophosphate deaminase 2 (AMPD2). This disorder is characterized by progressive atrophy of the cerebellum and brainstem, absent or small corpus callosum, and progressive postnatal microcephaly. Affected patients have severe global developmental delay often leading to death in early childhood [Akizu et al., 2013; Marsh et al., 2015]. AMPD2 deaminates adenosine monophosphate (AMP) to inosine monophosphate (IMP). Previous reports have attributed loss of function AMPD2 variants to adenosine-mediated neurotoxicity and defective protein synthesis secondary to guanine nucleotide deficiency as central to the pathogenesis of PCH9 [Akizu et al., 2013]. AMPD2 is one of three adenosine monophosphate deaminase paralogs. To date, all six reported PCH9-associated AMPD2 pathogenic variants have localized within the catalytic AMP deaminase domain, affecting all five predicted isoforms [Akizu et al., 2013; Marsh et al., 2015]. Here we report the clinical and genetic analysis of an individual with PCH9 secondary to a novel missense variant with strong evidence of pathogenicity, located outside the catalytic domain of AMPD2.
The family is of Middle Eastern origin. The parents are first cousins and unaffected. The male affected offspring showed clinical and brain imaging features consistent with PCH9 (Online Supplemental Table SI). He was born at term by normal vaginal delivery following an unremarkable pregnancy. He was well at birth and in the immediate neonatal period. At birth his OFC was 35.5 cm (40th centile) and weight was 3,850 g (80th centile). From early infancy he had significant global developmental delay and was diagnosed with cerebral palsy by his pediatrician. When first seen by a child neurologist at age 11 months his OFC was 43 cm, 0.5 cm <3rd centile. He was sleepy and was unable to fix or follow. He had marked axial hypotonia with head lag, and appendicular hypertonia with brisk reflexes and up-going plantar responses. A brain MRI scan performed at age 12 months showed absence of the corpus callosum, an interhemispheric cyst, and pontocerebellar hypoplasia (Fig. 1A). Extensive investigations, including biochemical, and metabolic studies, failed to identify a cause for his clinical presentation. Nerve conduction studies at age 12 months were normal. Over the next 14 months he progressively deteriorated, with increasing spasticity, decreased responsiveness, and an inability to feed orally necessitating insertion of a nasogastric tube. Two days following percutaneous insertion of a gastrostomy tube he deteriorated with pneumonia. A palliative approach was taken and he died from pneumonia at age 26 months.
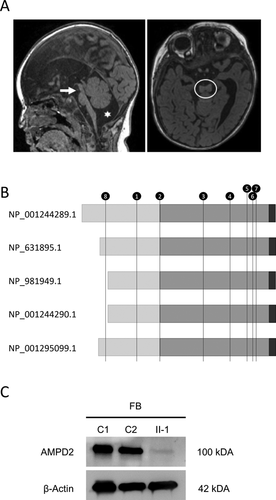
Chromosome microarray did not identify any pathological structural abnormalities but did show several long stretches of homozygosity, including the region spanning AMPD2 (Online Supplemental Table SII). Exome sequencing (ES) was performed by the Melbourne Genomics Health Alliance (MGHA, Human Research Ethics Committee approval 13/MH/3260). Prospective approval for further study was provided by the Royal Children's Hospital Research Ethics Committee (Human Research Ethics Committee #29077), with informed consent provided by participants or their guardians. The ES and variant analyses utilizing the MGHA shared bioinformatics pipeline [Sadedin et al., 2015; Stark et al., 2016] identified a novel, homozygous likely pathogenic AMPD2 variant. The variant (NM_001257360.1:c.751C>T, p.[Arg251Trp]), located within an exon present in all five mRNA isoforms, was verified by Sanger sequencing and both parents were confirmed to be carriers. Despite its location outside the catalytic AMP deaminase domain (Fig. 1B), the arginine residue is conserved, and the missense variant predicted to be damaging by PolyPhen-2, SIFT, and SNAP2. The ES did not identify any potential damaging variants in other known PCH genes (Online Supplemental Table SIII). Western blot analysis employing a monoclonal antibody (HPA036471, Sigma–Aldrich) directed against AMPD2 identified a single band, corresponding to the expected size of the predominant protein isoform, in protein extracts derived from control fibroblast cells. However, protein from patient-derived fibroblasts showed severe reduction in steady-state levels of AMPD2 (Fig. 1C).
AMPD2 is a critical component of the de novo purine synthetic pathway in humans and pathogenic variants leading to loss of protein function result in the severe neurodegenerative disorder PCH9 (OMIM#615809). Like other proteins associated with PCH, the function of AMPD2 has been shown to be critical for protein synthesis and energy homeostasis and highlights the unique susceptibility of neural cells to metabolic imbalances, both in a pre- and postnatal-setting [Paushkin et al., 2004; Namavar et al., 2011]. Here we identified a novel homozygous missense variant in AMPD2 as the molecular basis for disease in an individual presenting with clinical features consistent with PCH9 (Online Supplemental Table SI) [Akizu et al., 2013; Marsh et al., 2015]. There was no evidence of peripheral axonal neuropathy when tested at 12 months of age, consistent with the suggestion that this feature is age-dependent [Marsh et al., 2015]. Unlike previously reported individuals with PCH9, the pathogenic variant in the affected individual is located outside the catalytic AMP deaminase domain of the protein. Our results suggest the missense variant destabilizes AMPD2 and causes rapid degradation of the mutant protein. AMPD2 is one of three adenosine monophosphate deaminase (AMPD) orthologs responsible for the deamination of AMP to IMP as part of the de novo purine synthetic pathway in mammals. Human AMPD2 isoforms possess conserved C-termini but divergent N-termini, due to the use of alternative splicing and multiple promoters. These isoenzymes demonstrate unique expression profiles and catalytic behaviors [Haas and Sabina, 2003a,2003b] and appear to be associated with variable disease presentation. For instance, the likely pathogenic (c.319delT, p.[Cys107Alafs*80]) truncating variant, which is predicted to disrupt only three of five AMPD2 isoforms, was reported to cause the relatively mild disorder hereditary spastic paraplegia type 63 (OMIM#615686) [Novarino et al., 2014]. Affected individuals had normal cognition and no brainstem or cerebellar signs or peripheral neuropathy [Novarino et al., 2014]. The combined complexity of AMPD homolog expression and AMPD2 isoform production may underpin the vulnerability of certain brain structures in PCH9. For example, individuals with PCH9 have severe neurodegeneration observed in the cerebellum compared to the cerebral cortex. Previous work showed that AMPD2 is the predominant AMPD ortholog expressed in the cerebellum, whereas both AMPD2 and AMPD3 are coexpressed in the cerebral cortex [Akizu et al., 2013]. In addition, distinct regional demands for guanine and adenosine triphosphates during development may also contribute to the specific vulnerabilities of certain brain structures to neurodegeneration in PCH9. Future studies are required to determine the functional redundancy of AMPD2 and AMPD paralogs to understand the contribution of each to human disease development and presentation.
This is the first report linking PCH9 to mutation of AMPD2 outside the conserved AMP deaminase domain, the functional catalytic domain of the protein required for deamination of AMP to IMP as part of the de novo purine synthetic pathway. The variant is predicted to disrupt all five AMPD2 isoforms and like all previously described pathogenic missense variants results in significantly reduced steady state levels of AMPD2. The clinical features and disease progression are similar to previously described individuals with PCH9 resulting from pathogenic variants within the catalytic domain.
ACKNOWLEDGMENTS
The authors thank the family for participating in this study and are grateful for the generous support of the Lefroy and Handbury families. They thank Greta Gillies (MCRI) for assistance with sample preparation. This work was funded in part by National Health and Medical Research Council (NHMRC) Australia Project Grant (GNT1059666). APLM was supported by an Australian Postgraduate Award, RJL was supported by a Melbourne Childrens Clinician Scientist Fellowship and PJL was supported by a NHMRC Career Development Fellowship (GNT1032364). This work was supported by the Victorian Government's Operational Infrastructure Support Program and Australian Government NHMRC IRIISS.