Whole exome sequencing identifies the first STRADA point mutation in a patient with polyhydramnios, megalencephaly, and symptomatic epilepsy syndrome (PMSE)
Abstract
Polyhydramnios, megalencephaly, and symptomatic epilepsy syndrome (PMSE) is an ultra rare neurodevelopmental disorder characterized by severe, infantile-onset intractable epilepsy, neurocognitive delay, macrocephaly, and craniofacial dysmorphism. The molecular diagnosis of this condition has thus far only been made in 16 Old Order Mennonite patients carrying a homozygous 7 kb founder deletion of exons 9–13 of STRADA. We performed clinical whole exome sequencing (WES) on a 4-year-old Indian male with global developmental delay, history of failure to thrive, infantile spasms, repetitive behaviors, hypotonia, low muscle mass, marked joint laxity, and dysmorphic facial features including tall forehead, long face, arched eyebrows, small chin, wide mouth, and tented upper lip. A homozygous single nucleotide duplication, c.842dupA (p.D281fs), in exon 10 of STRADA was identified. Sanger sequencing confirmed the mutation in the individual and identified both parents as carriers. In light of the molecular discoveries, the patient's clinical phenotype was considered to be a good fit for PMSE. We identified for the first time a homozygous point mutation in STRADA causing PMSE. Additional bi-allelic mutations related to PMSE thus far have not been observed in Baylor ∼6,000 consecutive clinical WES cases, supporting the rarity of this disorder. Our findings may have treatment implications for the patient since previous studies have shown rapamycin as a potential therapeutic agent for the seizures and cognitive problems in PMSE patients. © 2016 Wiley Periodicals, Inc.
INTRODUCTION
Whole-exome sequencing (WES) using massively parallel next-generation-sequencing has proven to be powerful for molecular diagnosis of ultra rare diseases [Yang et al., 2013, 2014]. Polyhydramnios, megalencephaly, and symptomatic epilepsy syndrome (PMSE, MIM# 611087) is a neurodevelopmental disorder that has thus far only been reported in 16 Old Order Mennonite patients with a homozygous 7 kb founder deletion in STRADA (MIM# 608626, STE20-related kinase adaptor α, a.k.a. LYK5) [Puffenberger et al., 2007]. STRADA encodes STE20-related kinase adaptor protein, an upstream inhibitor of mammalian target of rapamycin complex 1 (mTORC1). Depletion of STRADA causes constitutive activation of the mammalian target of rapamycin (mTOR) signaling pathway. Here we report the utility of WES as the diagnostic tool for a consanguineous family from India with a child affected with PMSE and harboring a homozygous truncating mutation in STRADA.
CLINICAL REPORT
The proband was a 5-year-old male of Indian descent. He was born to a 26-year-old G1P1 mother and 28-year-old father. Prenatal ultrasounds showed polyhydramnios emerging as early as 12 weeks. The patient was delivered at full term via cesarean section for failure to progress with a birth weight of 2,306 g (1st percentile), birth length 49 cm (32nd percentile), and head circumference 33.5 cm (22nd percentile).
He was hospitalized at 10 days of age for feeding difficulties and poor weight gain. At 3 months of age, he presented with seizures, which were characterized as shivering, straightening of the legs, and sometimes whole body shaking with deviation of the eyes. His seizures were initially controlled with topiramate but worsened at 6 months. He was diagnosed with infantile spasms and treated with a 1-month course of adrenocorticotropic hormone (ACTH). In addition, he was started on pyridoxine, biotin, and phenobarbital and had no further seizures. These were continued until age 3.5 years when he was re-evaluated. Oxcarbazepine was initiated, and he was weaned off pyridoxine, biotin, and phenobarbital. Currently his seizures are well controlled.
The patient has had global developmental delay since early infancy accompanied by severe hypotonia and marked joint laxity in both large and small joints. He was noted to have a subluxing right knee since birth and consequently wears a knee brace. Delayed motor milestones include sitting at 2 years and 8 months and crawling around 5 years. At his current age of 5 years, he can cruise using a walker. He remains nonverbal aside from guttural sounds, and his receptive language is greatly limited. He can clap his hands, shake his head, and indicate a few needs by limb gestures. No regression has been observed.
Family history is significant for parents being first cousins, originating from East India. A younger female sibling is healthy. There are no other similarly affected individuals in the family.
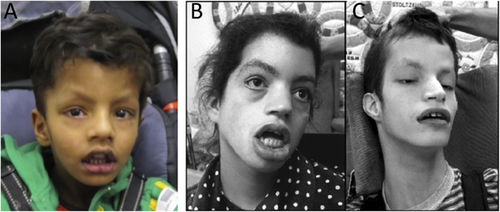
Physical examination at 5 years of age showed weight at the 22nd percentile, length at the 40th percentile, and head circumference at the 75th percentile. He had a thin body habitus with decreased muscle mass and minimal subcutaneous fat. Facial features included a tall forehead, narrow face, highly arched eyebrows, wide mouth, tented upper lip, and hypotonic facies with open mouth and drooling (Fig. 1). He had generalized marked joint laxity, and his right knee dislocated with minimal movement. He had generalized severe hypotonia. He did not have clonus or spasticity. Deep tendon reflexes were quiet but symmetric throughout. He made vocalizations but had no intelligible speech.
Diagnostic evaluations were all normal and included urine organic acid analysis, plasma amino acid profile, acylcarnitine profile, alpha-aminoadipic semialdehyde, urine creatine and guanidinoacetate, urine sulfocysteine, urine purine and pyrimidine profile, carbohydrate deficient transferrin, urine mucopolysaccharides, plasma copper, ceruloplasmin, lactic acid, pyruvate, and creatine kinase. A SNP chromosomal microarray identified long stretches of homozygosity involving many chromosomes and comprising 266 Mb, approximately 9.3% of the autosomal genome, consistent with parental consanguinity. No clinically significant deletions or duplications were detected. Brain magnetic resonance imaging (MRI) at 2 years 10 months showed only non-specific findings of a mild asymmetric decrease in the right periventricular parieto-occipital white matter substance with a small amount of somewhat linear abnormal periventricular white matter signal. The MR spectroscopy of the brain was normal. An electroencephalography (EEG) at 4 years of age showed multifocal epileptiform discharges (right frontal, left, and right central), and generalized background slowing. A renal ultrasound at 4 years 7 months showed medullary nephrocalcinosis, bilateral pelviectasis, and normal renal morphology and size. An echocardiogram at 4 years 8 months showed mesocardia, normal left and right ventricular size and systolic function, and normal valvular structure and function.
METHODS
The proband was referred to the Medical Genetics Laboratories at Baylor College of Medicine (BCM), Houston, TX, for whole exome sequencing (WES) analysis. Parental blood samples were received at the same time. Genomic DNA was extracted from peripheral whole blood. WES was performed on the proband and the data were analyzed as described previously [Yang et al., 2013, 2014]. Briefly, exome was captured using a solution-based exome capture reagent VCRome, version 2.1 (Roche NimbleGen, Madison, WI), and paired-end sequencing of 100-bp was performed on Illumina HiSeq 2,000 platform. For the proband, 8.6 Gb of uniquely aligned sequence was generated with an average coverage of 111 times for the targeted region, and 95.13% of targeted coding bases of the exome were covered by at least 20 reads. Preliminary filtering removed synonymous variants, intronic variants (over ± 5 bp away from an exon) and common benign variants with a MAF >1% in the exome sequencing project (ESP) 5,400 exomes or the 1,000 genomes project control databases, or >2% in Baylor internal database.
Illumina HumanExome-12v1 (cSNP) array analysis was run concurrently as a quality control measure. cSNP array contains >250k SNP markers in exonic regions. WES SNP genotype has a >99% concordance with cSNP array genotype. The array data were analyzed using GenomeStudio software (Illumina Inc., San Diego, CA), and cnvPartition 2.3.4 was used for detection of copy number variants and absence of heterozygosity (AOH). Sanger sequencing analysis was performed on the proband and the parents for variants in genes associated with autosomal dominant or X-linked disorders that are likely to be related to the patient's phenotype to confirm the variants and to determine the inheritance.
A photograph of the patient was collected after obtaining informed consent for publication of the photograph in the medical literature.
RESULTS
WES identified 780 variants in protein coding genes that were predicted to have an effect on protein coding or RNA splicing. Of these variants, 17 variants were in genes related to the patient's phenotype, including one deleterious variant in STRADA and 16 variants of unknown clinical significance.
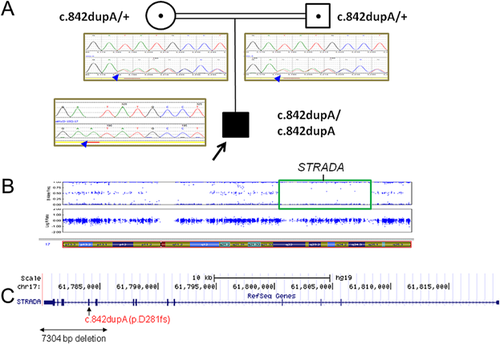
The deleterious STRADA variant is a homozygous single base pair duplication, NM_001003787:c.842dupA (p.D281fs) in exon 10 of STRADA. Duplication of a single nucleotide adenine at position c.842 is predicted to result in a translation frameshift with subsequent premature translation termination (Fig. 2). This variant was absent from the exome sequencing project (ESP), the 1,000 genomes project, and the Exome Aggregation Consortium (ExAC) databases. The homozygous pathogenic variant in STRADA was confirmed by Sanger sequencing in the proband. Sanger sequencing also showed that both parent are heterozygous for this mutation (Fig. 2A).
The other variants in disease genes related to the clinical phenotype in the proband included heterozygous missense or intronic changes in EHMT1, TSC2, KMT2A, and NALCN which are associated with autosomal dominant diseases, and a missense change in HUWE1 associated with an X-linked intellectual disability syndrome. None of these variants have been previously reported in patients. The variants in EHMT1, TSC2, and KMT2A were present in ExAC database, while variants in NALCN and HUWE1 were novel. Sanger sequencing confirmed these variants in the proband and showed that all of these variants were inherited from one parent with HUWE1 inherited from the mother.
cSNP array analysis revealed no evidence of large copy number changes in microdeletion/microduplication disease regions. However, continuous regions of copy neutral AOH greater than 5 Mb were detected on many chromosomes, consistent with previous SNP array results and the fact that the parents are first cousins. The STRADA mutation is located within one large AOH region on chromosome band 17q23.3 (Fig. 2B).
DISCUSSION
Loss of function mutations in STRADA cause PMSE, and the only pathogenic mutation reported thus far in STRADA is a 7 kb deletion of the last five exons, 9–13. Using WES, we identified for the first time a homozygous truncating point mutation in STRADA causing PMSE in a patient of Indian origin, thereafter correlating this result with the PMSE phenotype for its biological plausibility. Because of the non-specific nature of the PMSE phenotype and the fact that the molecular basis of the disorder had only been previously reported in the Old Order Mennonites, PMSE was not considered as a diagnosis during clinical evaluations. However, once the mutation was identified, it was felt that the patient's phenotype was consistent with PMSE.
In the 16 previously reported Mennonite patients, all had polyhydramnios, infantile-onset epilepsy, hypotonia, and craniofacial dysmorphism, as with our patient (Table I). The Mennonite patients had an age of seizure onset between 3 and 7 months concordant with our patient's seizure onset at 3 months. Seizures were frequent and medically refractory. Dysmorphic features in the Mennonite patients included a long face, prominent forehead, peaked eyebrows, broad nose, hypertelorism, and large mouth with thickened lips. Many of these features were present in our patient. Linear growth was normal in the Mennonite patients but body mass index (BMI) was consistently low (≤13 kg/m2) due to low muscle mass. Similarly, our patient had normal growth parameters and a BMI of 14 kg/m2. Interestingly, all but one of the Mennonite children had macrocephaly, some with a head circumference greater than four standard deviations above mean, but our patient had a normal head circumference, never exceeding the 75th percentile in 3 years of follow up. Neuroimaging in some of the Mennonite patients showed subependymal dysplasia, and ventriculomegaly contributing to macrocephaly. However, these features were absent in our case, suggesting modulation in the degree of activation of the mTOR pathway from modifying genes may play a role in the variability of macrocephaly. Alternatively, our proband's frameshift mutation in exon 10 of STRADA may engender lesser activation of the mTOR pathway than the STRADA deletion in the Mennonites, which could be tested through in vitro functional assays. The X-linked maternally inherited novel variant in HUWE1, which is associated with mental retardation, X-linked syndromic Turner type [MIM: 300706], may also contribute to the normal head circumstance in our case, as previously reported patients with HUWE1 mutations were either macrocephalic or normocephalic. Neurodevelopmental outcomes were uniformly poor in the Mennonite children. Gross motor skills were in the 6–14 month range, fine motor skills in the 4–10 month range, and language and communication skills in the 2–6 month range. Our patient had a similar, severe degree of psychomotor delay. Thirteen percent of the Mennonite patients had nephrocalcinosis, also seen in our patient. Other features that were not present in our patient include atrial septal defect, diabetes insipidus, supraventricular tachycardia, and leukemia.
| Features | Percentage of PMSE patients affected (N = 16) | This case |
|---|---|---|
| Polyhydramnios | 100 | + |
| Infantile spasms | 100 | + |
| Craniofacial dysmorphism | 100 | + |
| Hypotonia | 100 | + |
| Psychomotor retardation | 100 | + |
| Decreased muscle mass | 100 | + |
| Nephrocalcinosis | 13 (2/16) | + |
| Macrocephaly (hydrocephalus + megalencephaly) | 94 (15/16) | − |
| Strabismus | 56 (9/16) | − |
| Cardiac anomaly | Atrial septal defect, 25 (4/16) | Mesocardia |
| Marked joint laxity | Not noted | + |
Our patient exhibits a few novel features, specifically, mesocardia and marked joint laxity not fully attributable to his thin habitus and marked hypotonia. These findings expand the phenotypic spectrum of PMSE although a second genetic diagnosis accounting for these abnormalities is possible given parental consanguinity.
Our WES findings have potential treatment implications for the patient. Rapamycin, an mTOR inhibitor, has been shown to prevent constitutive mTORC1 signaling in fibroblasts from patients with PMSE, and treatment of five PMSE patients using Rapamycin showed no complications, a reduction in seizure frequency and an improvement in receptive language [Parker et al., 2013].
ACKNOWLEDGMENTS
We thank the patient's family for their collaboration and for allowing us to share their information. We also thank Dr. Ghayda Mirzaa for collaborative discussions.