Exome sequencing identifies a de novo frameshift mutation in the imprinted gene ZDBF2 in a sporadic patient with Nasopalpebral Lipoma-coloboma syndrome
Abstract
Nasopalpebral lipoma-coloboma syndrome (NPLCS, OMIM%167730) is an uncommon malformation entity with autosomal dominant inheritance characterized by the combination of nasopalpebral lipoma, colobomas in upper and lower eyelids, telecanthus, and maxillary hypoplasia. To date, no genetic defects have been associated with familial or sporadic NPLCS cases and the etiology of the disease remains unknown. In this work, the results of whole exome sequencing in a sporadic NPLCS patient are presented. Exome sequencing identified a de novo heterozygous frameshift dinucleotide insertion c.6245_6246 insTT (p.His2082fs*67) in ZDBF2 (zinc finger, DBF-type containing 2), a gene located at 2q33.3. This variant was absent in parental DNA, in a set of 300 ethnically matched controls, and in public exome variant databases. This is the first genetic variant identified in a NPLCS patient and evidence supporting the pathogenicity of the identified mutation is discussed. © 2016 Wiley Periodicals, Inc.
INTRODUCTION
Nasopalpebral lipoma-coloboma syndrome (NPLCS, OMIM%167730) is a rare malformation syndrome of autosomal dominant inheritance primarily characterized by nasopalpebral lipomas, upper and lower eyelid colobomas, telecanthus, and maxillary hypoplasia. Penchaszadeh et al. [1982] first described the disease in 1982 in a family from Venezuela comprising nine affected subjects in three consecutive generations. To date, two familial [Penchaszadeh et al., 1982; Akarsu and Sayli, 1991] and four sporadic [Bock-Kunz et al., 2000; Moreira Gonzalez and Jackson, 2003; Babu et al., 2011; Chacon-Camacho et al., 2013] NPLCS cases have been reported in the literature. Frequent additional clinical features include broad forehead, displaced, or aplastic lacrimal punctae, persistent epiphora, aberrant eyelashes, conjunctival hyperemia, corneal and lens opacities, and divergent strabismus [Penchaszadeh et al., 1982; Chacon-Camacho et al., 2013]. NPLCS has complete penetrance and a relatively homogeneous phenotype in both familial and sporadic cases.
The pathophysiology of the disorder is unknown and defects in migration of cells of neural crest or a dysplasia of adipose tissue were previously proposed to explain the aberrant craniofacial development observed in the syndrome [Penchaszadeh et al., 1982]. To date, a specific genetic alteration has not been associated with this Mendelian disorder.
In this work, we report the identification of a de novo truncating mutation in the ZDBF2 (zinc finger, DBF-type containing 2) gene in a patient with the NPLCS detected by whole exome sequencing (WES) and summarize evidence supporting pathogenicity of such mutation.
PATIENT AND METHODS
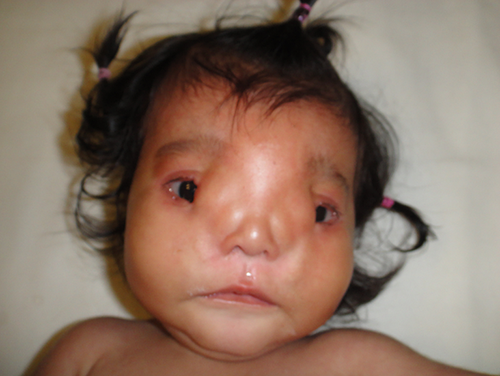
The propositus is a 2-year-old Mexican girl with a clinical diagnosis of NPLCS (Fig. 1). She was born from unrelated, healthy parents. No family history of congenital malformations or inherited diseases was recorded. The clinical, radiological, and histopathological features of this sporadic case were described by or group [Chacon-Camacho et al., 2013]. In an attempt to identify the genetic defect underlying the syndrome, samples from the proband, and her healthy parents were subjected to WES in the Baylor-Hopkins Centers for Mendelian Genomics (BHCMG).
Briefly, we captured the CCDS exonic regions and flanking intronic regions totaling ∼51 Mb by using the Agilent SureSelect Human All Exon V4 51 Mb Kit and performed paired end 100 bp reads on all three family members with the Illumina HiSeq2000 platform. We aligned each read to the reference genome (NCBI human genome assembly build 37; Ensembl core database release 50_36110) with the Burrows-Wheeler Alignment (BWA) tool [Li and Durbin, 2009] and identified single-nucleotide variants (SNVs) and small insertion-deletions (indels) with SAMtools [Li et al., 2009]. We also performed local realignment and base call quality recalibration by using GATK [McKenna et al., 2010; DePristo et al., 2011]. We identified potentially causal variants by standard filtering criteria: SNV and indel minimal depth of 83, root mean square mapping quality of 25, strand bias P-value below 104, end distance bias below 104, and filtering out SNVs within 3 bp of an indel and indels within 10 bp of each other; followed by the use of the Analysis Tool of PhenoDB [Sobreira et al., 2015] to design the prioritization strategy.
We prioritized rare functional variants (missense, nonsense, splice site variants, and indels) that were heterozygous de novo mutations, homozygous, or compound heterozygous in the proband and excluded variants with a Minor Allele Frequency (MAF) >0.01 in dbSNP 126, 129, and 131, in the Exome Variant Server (release ESP6500SI-V2), 1000 Genomes Project, or Exome Aggregation Consortium database (ExAC), Cambridge, MA (URL: http://exac.broadinstitute.org). We also excluded all variants found in our in-house controls (CIDRVar 51 Mb). Sanger sequencing confirmed candidate mutations Identified in WES. For distinguishing parental origin of the allele carrying the candidate causal mutation, allele-specific PCR employing primer ASO-RV: 5′-TGGTTGCTCCTATTAAAAAAT G- 3′ (underlined bases indicate a dinucleotide insertion) and Sanger sequencing were used. Complete oligonucleotide sequences and PCR conditions for allele-specific PCR are available on request.
RESULTS
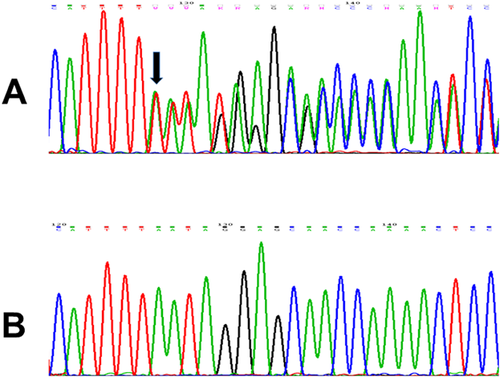
After filtering variants detected by WES, 1 heterozygous de novo variant, 12 compound heterozygous variants in six genes, and one homozygous variant were identified in propositus' DNA (Suppl. Table SI). The de novo variant is a novel heterozygous c.6245_6246insTT (p.His2082fs*67) insertion in the third coding exon of the ZDBF2 gene (NM_020923), which was absent from parental DNA on WES. Bidirectional Sanger sequencing confirmed that the proband carried the ZDBF2 dinucleotide insertion (Fig. 2) while her parents exhibited wild-type alleles. The variant has not been reported in the above-mentioned public variant databases and was absent from a set of 300 ethnically matched control alleles genotyped by direct Sanger sequencing. We also identified a second variant in ZDBF2 in the proband inherited from her mother (c.2086A>C; p.Asn696His) (Suppl. Table I), which was employed for recognition of the maternal ZDBF2 allele. As shown in supplementary Figure S1, allele-specific sequencing demonstrated that the ZDBF2 allele carrying the frameshifting dinucleotide insertion was the paternally derived one.
The remaining variants identified in proband's DNA by WES (i.e., a single homozygous mutation and six compound heterozygous mutations) were excluded by its presence in the parents, assuming a dominant mutation model for NPLCS.
DISCUSSION
The pathophysiology of the NPLCS is currently unknown. It has been suggested that the primary defect could involve a dysplasia of adipose tissue leading to nasopalpebral and upper lid lipomas during embryogenesis, with the rest of the malformations being secondary to interference of morphogenesis of the mid-upper face developmental field from the lipomatous hamartomas [Penchaszadeh et al., 1982]. Only two familial cases of the disease have been described and in both pedigrees the phenotype was transmitted as an autosomal dominant trait [Penchaszadeh et al., 1982; Akarsu and Sayli, 1991]. In this work, WES of a sporadic NPLCS case allowed the identification of a de novo truncating mutation in ZDBF2, a gene located on human chromosome 2q33.3 and composed of three coding exons that encodes a 2,354 amino acid polypeptide. Both ZDBF2 mRNA and protein are highly expressed in human tissues as prostate, cerebral cortex, adrenal gland, skin, and adipose tissue, among other body structures (www.proteinatlas.org/ENSG00000204186-ZDBF2/tissue). ZDBF2 is a nucleic acid binding protein with a DBF-zinc finger domain but its particular function is currently unknown.
Human ZDBF2 locus was first described to be expressed only from the paternal allele [Kobayashi et al., 2009]. However, recent work has shown that ZDBF2 is under atypical dynamic imprinting with both maternal and paternal imprinting control during development [Duffié et al., 2014]. ZDBF2 does not follow the rules of classic imprinted human genes that exhibit acquisition of a differentially methylated region in one of the parental germlines only and have an uninterrupted continuum of paternal or maternal-specific methylation from gametes to adult progeny [Kobayashi et al., 2012; Duffié et al., 2014].
Two lines of evidence support that the heterozygous p.His2082fs*67 truncating ZDBF2 variant identified in our patient is associated with her malformative phenotype. First, the heterozygous and truncating nature of the ZDBF2 variant identified in our patient (but not in her healthy parents) is compatible with an autosomal dominant inheritance fitting the transmission pattern described in familial cases of the disease. The variant was absent from a set of 300 ethnically matched control alleles and is not annotated in publicly available databases, including the recently released ExAC, which contains exome data of ∼4,000 Mexican people [SIGMA Type 2 Diabetes Consortium, 2014]. Second, loss of ZDBF2 in humans leads to a similar set of craniofacial malformations that includes some of the anomalies observed in the NPLCS. The DECIPHER database (https://decipher.sanger.ac.uk/) includes seven individuals with 2q33.3 deletions involving ZDBF2 and six of them feature craniofacial defects including abnormal palpebral fissure and/or eyelashes (IDs 249506, 249239, 249612, 256887, 292523) and prominent/wide nasal bridge (IDs 249506, 256887) (complete data available at https://decipher.sanger.ac.uk/search?q = ZDBF2#consented-patients/results).
It is important to make note that two instances of females transmitting NPLCS to their children have been described in familial cases of the disease [Penchaszadeh et al., 1982; Akarsu and Sayli, 1991], and this fact would invalidate the paternally active-ZDBF2 being the NPLCS causal gene. However, taking into account the recent findings demonstrating that ZDBF2 is under atypical dynamic imprinting with both maternal and paternal imprinting control during development [Duffié et al., 2014], the causality of ZDBF2 in such cases of female transmission cannot be excluded. Alternatively, the possibility of genetic heterogeneity, with mutations in a different gene causing the syndrome, can also be considered in those instances of NPLCS transmission from affected females.
In conclusion, we demonstrated a ZDBF2 mutation in a NPLCS patient and discussed evidence supporting the pathogenicity of this gene variant. However, genetic analyses of additional patients with this uncommon craniofacial malformation phenotype and/or the development of a knock-out model would provide additional evidence to confirm ZDBF2 involvement in the NPLCS.
ACKNOWLEDGMENTS
The authors thanks to the Baylor-Hopkins Center for Mendelian Genomics research initiative for whole exome sequencing and data analysis, and the grant from the National Human Genome Research Institute, (1U54HG006542) that provided support for this work.