Lateral meningocele (Lehman) syndrome: A child with a novel NOTCH3 mutation
Abstract
Lateral meningocele syndrome (LMS), or Lehman syndrome, is a rare disorder characterized by multiple lateral spinal meningoceles, distinctive facial features, joint hypermobility and hypotonia, along with skeletal, cardiac, and urogenital anomalies. Heterozygous NOTCH3 mutations affecting the terminal exon 33 were recently reported as causative in six families with LMS. We report a boy with LMS, the fourteenth reported case, with a de novo 80 base pair deletion in exon 33 of NOTCH3. Our patient's prenatal findings, complex cardiac anomalies, and severe feeding difficulties further expand our understanding of this rare condition. © 2016 Wiley Periodicals, Inc.
INTRODUCTION
Multiple lateral meningoceles are a rare anomaly. Lehman et al. [1977] first described a mother and daughter with similar craniofacial dysmorphisms, skeletal sclerosis, and multiple meningoceles. Philip et al. [1995] compared the phenotype of a 19-year-old man with multiple lateral meningoceles to Lehman's patients, and found similarities including malar hypoplasia, downslanting palpebral fissures, micrognathia, and skeletal anomalies. Gripp et al. [1997] described three unrelated patients with multiple lateral meningoceles and similar facial dysmorphism, and features of a connective tissue disorder with normal collagen studies. Chen et al. [2005] reported three additional patients, including an affected mother and daughter pair. Alves et al. [2013] reported the first case of a bicuspid aortic valve in a 5-year-old boy with multiple lateral meningoceles, further underscoring a possible connective tissue etiology; he was re-reported by Correia-Sa et al. [2015] to highlight Pierre-Robin sequence. Finally, Kumar et al. [2013] and Castori et al. [2014] described two adult patients with lateral meningoceles. An additional patient initially reported to have Hajdu–Cheney syndrome [Avela et al., 2011] was reclassified as lateral meningocele syndrome [Gripp, 2011; Gripp et al., 2015], for a total of 13 cases in eight families.
Lateral meningocele syndrome (LMS; OMIM 130720), also referred to as Lehman syndrome, encompasses characteristic facial features, lateral spinal meningoceles, hyperextensibility, hypotonia, as well as congenital heart defects, vertebral anomalies, abdominal hernias and cryptorchidism. Heterozygous mutations in the last exon of the NOTCH3 gene were recently shown to be associated with Lehman syndrome in six individuals [Gripp et al., 2015]. NOTCH3 is a part of an evolutionary conserved family of cell surface receptors that regulate gene expression and subsequently cell growth and proliferation [Baeten and Lilly, 2015].
We report the fourteenth case of Lehman syndrome and sixth identified with a NOTCH3 mutation, to further expand the genotype–phenotype correlation for this rare condition. Notably, the mutation, an 80 base pair (bp) deletion in exon 33, is novel.
CLINICAL REPORT
The patient was a 2 years and 8 months old boy born to a non-consanguineous Caucasian 42-year-old G3P2SA1 mother and 41-year-old father. Antenatal anatomy ultrasound showed nuchal edema, echogenic bowel, and an abnormal cardiac 4-chamber view. Fetal echocardiogram at 22 weeks showed hypoplasia of the aortic arch, transverse arch and isthmus, with a bicuspid aortic valve. He was born at term by induced vaginal delivery with vacuum assist due to fetal heart rate decelerations. Apgar scores were 8 at 1, 5, and 10 min. Prostaglandin infusion and non-invasive positive pressure ventilation were started in the delivery room. Birth weight was 3.4 kg, length was 51 cm, and occipitofrontal circumference (OFC) was 34 cm, all approximately 50th centile. Postnatal echocardiogram showed tubular hypoplasia of the aortic arch with coarctation, large ventricular septal defect, large ostium secundum type atrial septal defect, bicuspid aortic valve, and bilateral superior vena cavae.
During open repair of the cardiac anomalies at day 3 of life, he had junctional ectopic tachycardia and required extra-corporeal membrane oxygenation (ECMO). Post-operatively, head ultrasound showed right occipital cerebral hemorrhage. Magnetic resonance imaging (MRI) of the brain at 23 days of life showed Chiari I malformation with cerebellar tonsils extending to the posterior arch of C2 (Fig. 1A) and middle cranial fossa encephaloceles (Fig. 1B).
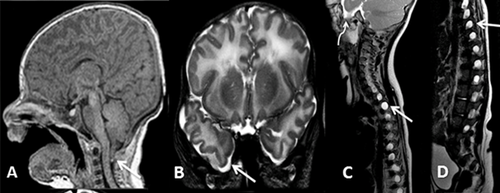
After discharge at 2 months of age, parents noted bilateral flank swelling. An ultrasound of the abdomen showed lateral lumbar meningoceles displacing the kidneys and causing the perceived swelling due to deficient posterior wall musculature. Spine MRI showed enlarged spinal canal and bilateral extensive lateral neural foramina meningoceles (Fig. 1C and D). The baby was discharged with nasogastric tube in situ for feeding, and eventually required gastrostomy tube for dysphagia. At age 3 months, he was diagnosed with moderate bilateral conductive hearing impairment; hearing aids were prescribed. He had left amblyopia likely secondary to right occipital hemorrhage. Cardiac status was stable. He had neck hyperextension and noisy breathing during sleep, but polysomnogram did not show airway obstruction. Prenatal karyotype, postnatal array comparative genomic hybridization (aCGH), and a Noonan syndrome genetic testing panel were normal.
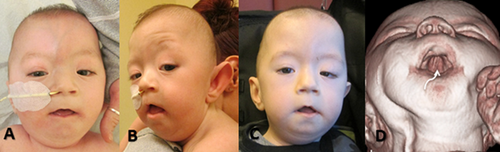
At 2 years and 8 months, weight was 12.2 kg (10–25th centile), height 92.6 cm (25–50th centile), and OFC 48.3 cm (−1 SD). Dysmorphic features included left posterior plagiocephaly and tall cranial vault, sparse hair, hypoplastic supraorbital ridges, epicanthus, hypertelorism, ptosis, downslanting palpebral fissures, midface hypoplasia, small nares, long philtrum, and thin upper vermillion, highly arched palate, bifid uvula, short upper lingual frenulum, asymmetric low set ears with short canals, microretrognathia, and bilateral single palmar creases (Fig. 2). He had a hoarse and weak cry. Neurological exam was significant for paucity of facial movement, diffuse hypotonia and joint hypermobility.
Psychomotor development was significantly delayed. On assessment at 13 months, he was beginning to tripod sit and was unable to stand independently. He had a palmar grasp. He was able to turn objects over in his hands and throw them. Expressive language was limited to “mama” and “dada” and he was able to wave and clap on command. At 2 years and 8 months, he had recently started to take independent steps with a wide based gait. Expressive language development had not progressed. Receptive language progressed to following one-step commands such as blowing kisses. He was able to shake his head to gesture “no.”
Family history was negative for connective tissue disease and developmental disorders. Neither parent had observable clinical features of LMS.
The family was enrolled in the Care4Rare Canada research project to identify the gene for LMS. Institutional research ethics board approval was obtained (Hospital for Sick Children and Children's Hospital of Eastern Ontario) and the trio was enrolled with informed consent. DNA was extracted by standard protocols. Whole-exome sequencing (WES) was performed on genomic DNA from the proband and his unaffected parents. Agilent SureSelect Human All Exon Kit version 5 was used for target enrichment, and the library was sequenced using 100 base pair paired-end runs on an Illumina HiSeq 2,000 platform at McGill University and Genome Quebec Innovation Centre (Montreal, Canada). The bioinformatics analysis was carried out as previously described [Tetreault et al., 2015], using BWA for mapping of reads against the human reference genome (hg19), Samtools mpileup for variant calling and a combination of Annovar and custom scripts for variant annotation. A mean coverage of 147X (patient) and 135X (both parents) was obtained. Approximately 97% of the bases were covered by ≥10 reads as determined by the Genome Analysis Toolkit. CNV analysis using coverage depth comparison was performed using FishingCNV [Shi and Majewski, 2013]. The WES data from the trio was analyzed for de novo mutations but no compelling candidate was identified.
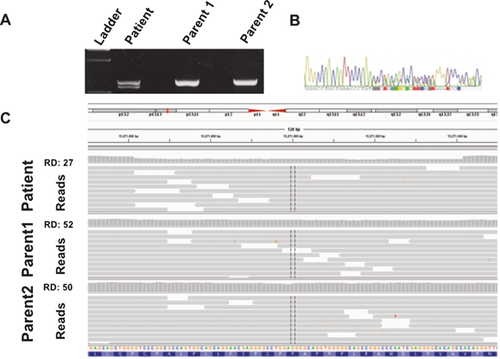
Upon publication of Gripp et al.'s [2015] report linking mutations in exon 33 of NOTCH3 to LMS, we reanalyzed the WES data from our patient but did not identify de novo rare variants in exon 33 of NOTCH3. We then PCR-amplified and sequenced this region (Fig. 3A and B); an 80 base pair deletion in NOTCH3 was identified (NM_000435.2:c6498_6577del; p.Ala2167Profs*48), resulting in a premature stop codon in exon 33. Subsequent analysis of the parents demonstrated that it was a de novo change. Manual inspection of the WES data from this region of NOTCH3 in the patient using Integrative Genomics Viewer (https://www.broadinstitute.org/igv/) identified the deletion (Fig. 3C).
DISCUSSION
Our patient is the 14th reported individual with LMS, with a de novo previously unreported heterozygous NOTCH3 mutation. Gripp et al. [2015] recently reported five de novo truncating NOTCH3 mutations in six unrelated individuals, all within the last exon of the gene, exon 33. Our patient has the largest reported deletion causing truncation of exon 33. One of Gripp et al.'s [2015] reported patients had a 25 base pair deletion in NOTCH3. For our patient, the mutation was missed by both standard data analysis pipeline, which would pick up a maximum of 30 base pair deletions, and by fishing CNV. Our experience demonstrates that due to limitations of current analysis pipeline methods, WES does not comprehensively allow for mutation discovery, nor does it replace targeted sequencing.
Mutations in this last exon of NOTCH3 are predicted to damage or omit the PEST domain, which is located in the terminal exon of all Notch family genes, and promotes degradation of the intracellular signaling portion of the activated protein via proteasomes [Wang et al., 2015]. Thus, truncating mutations in this region may prolong Notch3 signaling by increasing its cellular half-life [Gripp et al., 2015]. Interestingly, mutations in the final exon of NOTCH2, which is co-expressed with NOTCH3 on vascular smooth muscle endothelium, cause Hajdu–Cheney syndrome (OMIM 102500), a condition with overlapping clinical characteristics [Majewski et al., 2011].
Our patient's phenotype appears to be at the severe end of the spectrum based on previous reports. In Table I, we compare his phenotype with the six reported patients with NOTCH3 mutations. The severity of his phenotype is most notable in feeding difficulties necessitating enteral feeding, developmental delay affecting language development as well as the expected motor delays, and the most complex cardiac anomalies reported in LMS. Confounding factors include a prolonged hospital course and surgical complications requiring ECMO, and the resultant right occipital hemorrhage, which may contribute to delay in meeting developmental milestones [Hanekamp et al., 2006]. Table I also suggests that screening for hearing impairment, Chiari 1 malformation, and cardiovascular malformations, particularly aortic abnormalities, should be considered in patients in whom the diagnosis of LMS is suspected.
| LMS patients with NOTCH3 mutations [Gripp et al., 2015] | |||||||
|---|---|---|---|---|---|---|---|
| Features | Our patient | ID 1 | ID 7 | ID 15 | ID 20 | ID 26 | ID 28 |
| Sex | Male | Male | Male | Male | Male | Male | Male |
| Age (years) | 2 | 24 | 10 | 8 (age at death) | 10 | 9 | 5 |
| NOTCH 3 mutation | c.6498-6577del | c.6461-6486del | c.6692_93insC | c.6692_93insC | c.6732C>A | c.6663C>G | c.62474A>T |
| Inheritance | De novo | De novo | N/A | De novo | De novo | De novo | De novo |
| Prenatal abnormalities | Increased NT, echogenic bowel, cardiac anomalies | − | N/A | − | − | Cystic hygroma | − |
| Developmental Delay | + | + | + | + | + | + | + |
| Intellectual Disability | N/A | + | − | N/A | − | − | − |
| Hypotonia | + | + | + | + | + | + | + |
| Hyperextensibility | + | + | + | + | + | + | + |
| Hearing impairment | + conductive | − | − | − | + mixed | + conductive | − |
| Brain anomalies | Chiari 1, middle cranial fossa encephaloceles | Mild ventricular dilatation | N/A | Chiari 1, hydrocephalus | − | − | N/A |
| Eye anomalies | Left amblyopia | − | − | − | − | Bilateral upward gaze restriction | − |
| Cardiac anomalies | Tubular hypoplasia of the aortic arch with coarctation, VSD, ASD, bicuspid aortic valve, bilateral SVC | Aortic dilation | − | VSD, aberrant right subclavian artery, interrupted inferior vena cava | VSD, aortic dilation | Biscupid aortic valve | − |
| Feeding difficulties | Dysphagia (G-tube fed) | − | − | Dysphagia | GERD, poor feeding | GERD | Slow weight gain |
- +, feature present; −, feature absent; N/A, not available; VSD, ventricular septal defect; ASD, atrial septal defect; SVC, superior vena cava; GERD, gastroesophageal reflux disease.
Congenital cardiovascular malformations have been described in four of fourteen reported patients; all four patients had NOTCH3 mutations. Bicuspid aortic valve was noted in one patient and dilatation of the aorta was noted in two [Alves et al., 2013; Gripp et al., 2015]. Our patient presented prenatally with more extensive cardiac anomalies requiring surgical repair. The NOTCH3 gene may play a pro-proliferative role in aortic and vascular smooth muscles [Baeten and Lilly, 2015]. Mutations in other regions of NOTCH3 cause CADASIL, a cerebral arteriopathy, and may contribute to some cases of childhood-onset pulmonary arterial hypertension [Chida et al., 2014], suggesting that NOTCH3 signaling has an important role in the development and/or maintenance of vascular tissue.
Similarities between the facial features of individuals with LMS and Noonan syndrome and related RASopathies have been previously observed [Gripp et al., 2015], including hypertelorism, ptosis, epicanthal folds, low-set, and posteriorly-angulated ears, and a low posterior hairline. Our patient had prenatal genetic testing for RASopathies due to nuchal edema and congenital heart anomalies. Similar to the Ras/MAPK pathway, Notch3 acts as a pro-proliferative agent [Baeten and Lilly, 2015]. Communication between the MAPK/ERK pathway and Notch3 has been previously established [Haruki et al., 2005] and Baeten and Lilly [2015] recently showed the Notch3 acts through the MAPK/ERK pathway to upregulate downstream pro-proliferative genes. Thus, LMS should be considered in the differential diagnosis of prenatal and infantile Noonan syndrome-like findings with negative molecular testing.
In conclusion, we report the sixth case of NOTCH3 mutation-positive LMS/Lehman syndrome. Further characterization of human phenotypes associated with the NOTCH gene family mutations will enhance our understanding of the molecular pathogenesis of aberrant NOTCH signaling.
ACKNOWLEDGMENTS
We thank our patient and his family for their participation. This disorder was selected for study by the Care4Rare Canada (Enhanced Care for Rare Genetic Diseases in Canada) Consortium Gene Discovery Steering Committee: Kym Boycott (lead; University of Ottawa), Alex MacKenzie (co-lead; University of Ottawa), Jacek Majewski (McGill University), Michael Brudno (University of Toronto), Dennis Bulman (University of Ottawa), and David Dyment (University of Ottawa) and funded, in part, by Genome Canada, the Canadian Institutes of Health Research, the Ontario Genomics Institute, Ontario Research Fund, Genome Quebec, Children's Hospital of Eastern Ontario Foundation and the Hospital for Sick Children. The authors wish to acknowledge the contribution of the high throughput sequencing platform of the McGill University and Génome Québec Innovation Centre, Montréal, Canada.