Novel missense mutations in a conserved loop between ERCC6 (CSB) helicase motifs V and VI: Insights into Cockayne syndrome
Abstract
Cockayne syndrome is caused by biallelic ERCC8 (CSA) or ERCC6 (CSB) mutations and is characterized by growth restriction, microcephaly, developmental delay, and premature pathological aging. Typically affected patients also have dermal photosensitivity. Although Cockayne syndrome is considered a DNA repair disorder, patients with UV-sensitive syndrome, with ERCC8 (CSA) or ERCC6 (CSB) mutations have indistinguishable DNA repair defects, but none of the extradermal features of Cockayne syndrome. We report novel missense mutations affecting a conserved loop in the ERCC6 (CSB) protein, associated with the Cockayne syndrome phenotype. Indeed, the amino acid sequence of this loop is more highly conserved than the adjacent helicase motifs V and VI, suggesting that this is a crucial structural component of the SWI/SNF family of proteins, to which ERCC6 (CSB) belongs. These comprise two RecA-like domains, separated by an interdomain linker, which interact through helicase motif VI. As the observed mutations are likely to act through destabilizing the tertiary protein structure, this prompted us to re-evaluate ERCC6 (CSB) mutation data in relation to the structure of SWI/SNF proteins. Our analysis suggests that antimorphic mutations cause Cockayne syndrome and that biallelic interdomain linker deletions produce more severe phenotypes. Based on our observations, we propose that further investigation of the pathogenic mechanisms underlying Cockayne syndrome should focus on the effect of antimorphic rather than null ERCC6 (CSB) mutations. © 2016 Wiley Periodicals, Inc.
INTRODUCTION
Cockayne syndrome (CS; OMIM 133540/216400) is a rare, autosomal recessive disorder characterized by restricted postnatal growth, disproportionate microcephaly, developmental delay, and premature pathological ageing [Wilson et al., 2015]. It is principally caused by mutations in ERCC6 (CSB; OMIM 609413) and ERCC8 (CSA; OMIM 609412) and is historically considered a DNA repair disorder [Horibata et al., 2004; Nardo et al., 2009]. ERCC8 (CSA) and ERCC6 (CSB) cooperate in the recognition of bulky DNA adducts during transcription. ERCC6 (CSB) is a nuclear resident protein, which recognizes and binds such lesions in concert with XPG [Sarker et al., 2005]. ERCC8 (CSA) is a cytoplasmic E3-ubiquitin ligase, which is recruited into the nucleus in response to DNA damage in complex with its chaperone, UVSS-A [Saijo et al., 2007]. ERCC8 (CSA) polyubiquitinates ERCC6 (CSB), removing this protein from damaged template strand lesions, which are then resolved through the common nucleotide excision repair pathway. The fibroblasts from patients with classical CS exhibit a defect in transcription-coupled nucleotide excision repair (tc-NER) following UVC irradiation. However, defective tc-NER cannot be the cause of the CS phenotype, as an indistinguishable DNA repair defect caused by mutations in ERCC8 (CSA) or ERCC6 (CSB) occurs in patients with UV-sensitive syndrome (UVSS; OMIM 600630/614621), who experience photosensitivity, but lack the impaired growth, developmental problems, or signs of pathological ageing seen in patients with CS. A number of alternative roles for the ERCC6 (CSB) protein have been proposed, including altering the action of base excision repair factors [Wong et al., 2007], in telomere maintenance [Batenburg et al., 2012] and in mitochondrial DNA repair [Scheibye-Knudsen et al., 2012]. However, ERCC8 (CSA) has not been found to participate in any of these processes, suggesting that while defects in these pathways may act as modifiers, they are not the major influence on the CS phenotype. Further clues are needed to establish the pathology underlying CS. Here, we describe two patients from different ethnic backgrounds with novel homozygous ERCC6 (CSB) mutations in an evolutionarily conserved motif common to the SWI/SNF family of proteins, associated with the CS phenotype.
CLINICAL REPORTS
Patient 1 (Fig. 1A) is the first child of consanguineous parents. He was born at term, weighing 3.3 kg (–0.5 SD). He presented at 2.5 years with postnatal growth failure, disproportionate microcephaly, and developmental delay. He developed intention tremor at 5 years and had a broad-based ataxic gait. He also had a mild, predominantly motor demyelinating polyneuropathy and proximal weakness of the legs. He had bilateral sensorineural hearing loss from 15 years. He was hypermetropic and underwent regular ophthalmology evaluations, but did not have cataracts or retinal abnormalities. He had subcutaneous fat loss, although the age at onset was unclear. He also had a photosensitive rash, managed with the use of sunscreen. At 22 years, his height was 125 cm (–7.7 SD), weight was 25 kg (–12.6 SD), and OFC was 49 cm (–4.8 SD for 18 y). He was initially diagnosed with CS following skin biopsy, as his fibroblasts demonstrated impaired recovery of RNA synthesis after UVC irradiation (Fig. 1B).
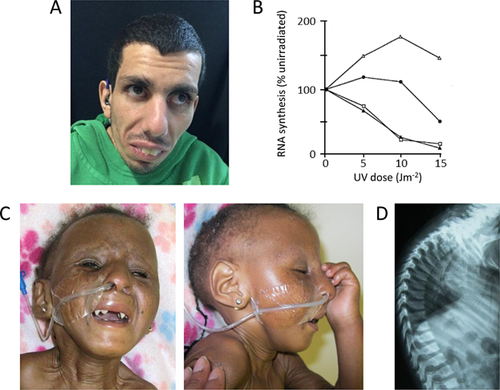
Patient 2 (Fig. 1C) is the second child of South African parents who are not known to be consanguineous, but belong to the same tribe. She was born at term, weighing 2.1 kg (–3.1 SD) with an OFC of 33 cm (–1.3 SD). She had postnatal growth failure with disproportionate microcephaly. Following normal early development, she regressed from 8 months of age. She had a single febrile seizure at 10 months of age. She had bilateral divergent strabismus and progressive eye abnormalities, comprising bilateral corneal clouding and nystagmus. She also had bilateral conductive hearing loss. Dental anomalies included conical teeth and multiple dental caries, likely representing enamel hypoplasia. She also had subcutaneous fat loss with loose skin, and developed progressive contractures, principally affecting her large joints. Additionally, she had a severe thoracic kyphosis (Fig. 1D). She died at 4 years of age.
METHODS
Both patients were recruited to the Cockayne Syndrome Natural History study, which was granted ethics approval by the NRES Committee North East—Newcastle and North Tyneside 2. Written consent was obtained from both families for participation and publication of clinical information. For Patient 1, recovery of RNA synthesis in fibroblasts following UVC irradiation was assessed as described [Colella et al., 1999], before recruitment to our study. Genomic DNA extracted from blood was used for genetic investigations. Bidirectional fluorescent Sanger sequencing was performed on amplicons encompassing all exons of ERCC6, including non-coding exon 1, and flanking intronic sequences; data were analyzed using the current version of Mutation Surveyor™ software. Details of primers are available on request.
RESULTS
Sanger sequencing of the ERCC8 (CSA) and ERCC6 (CSB) genes showed patient 1 to be homozygous for ERCC6 (CSB) c.2800C>A, p.Pro934Thr (Supplemental Online Fig. S1A); both parents were confirmed to be carriers. Patient 2 was apparently homozygous for ERCC6 (CSB) c.2808G>C, p.Trp936Cys (Supplemental Online Fig. S1B). In this case, testing of both parents was not possible to rule out the presence of a deletion in trans with the missense mutation. Further samples were not available from the proband for quantitative PCR or MLPA. However, as Patient 2 was heterozygous for other SNPs in ERCC6 (CSB), including in the same fragment as the reported missense mutation, and both parents belong to the same tribe, it is probable that this change is homozygous.
DISCUSSION
Pro934 and Trp936 form part of a loop between SWI/SNF helicase motifs V and VI. While much attention has been paid to the adjacent motifs, the role of this loop has not been evaluated previously. Throughout this family of proteins, this loop has retained an identical amino acid sequence from soil living amoeba (slime mould), through to man (Fig. 2A). Such remarkable conservation suggests that this structure is essential for the function of SWI/SNF proteins generally. Crystal structures show that SWI/SNF family members, such as Saccharomyces cerevisae Rad54, adopt a bilobar structure, with two RecA-like domains [Thomä et al., 2005]. Helicase motifs I, IA, II, and III contribute to the N-terminal RecA-like domain, while motifs IV, V, and VI form the C-terminal domain. A linker region between motifs III and IV allows the two Rec-A-like domains to interact, forming an interdomain cleft. Motif VI is essential for this interdomain association. As this conserved loop lies buried close to this cleft and is immediately upstream of motif VI (Fig. 2B), it may be that mutations here affect the ability of motif VI to associate with the first RecA-like domain, disrupting the tertiary protein structure.
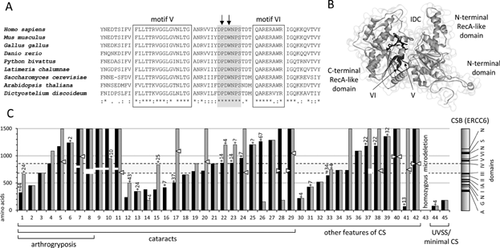
In an attempt to glean further mechanistic insights from available ERCC6 (CSB) mutation data, we reanalyzed a recent review of published mutations in CS and cerebro-oculofacioskeletal syndrome [Laugel et al., 2010], which is essentially a prenatal onset form of the disorder. What becomes clear from this analysis is that homozygous or compound heterozygous missense mutations are uncommon molecular causes of CS (5 of 44 allele combinations, including the patients reported here; Fig. 2C). Most patients have at least one truncating mutation (nonsense/frameshift). The next commonest change in ERCC6 (CSB) is deletion of residues from the interdomain linker between motifs III and IV, which tends to be seen in patients with more severe phenotypes. Interestingly, most amino acid substitutions caused by missense mutations also lie within this region. There is no association between the length of truncated ERCC6 (CSB) and phenotype, except that if both alleles predict proteins <200 amino acids, the patient displays minimal features of CS, or a UVSS phenotype. Mutations that result in major disruption of the tertiary structure of ERCC6 (CSB), for example, truncations and changes preventing physical interaction of the two RecA-like domains, can be termed antimorphic. These data suggest that in order to develop CS, a patient generally requires two antimorphic mutations, or one antimorphic mutation in tandem with a hypomorphic allele. The mutations we report here in the conserved motif V–VI loop are in keeping with this, as putative structural determinants of ERCC6 (CSB) interdomain stability. The principal mechanism underlying CS pathogenesis is currently unknown. We hypothesize that in patients with ERCC6 (CSB) mutations, this mechanism will be dependent on the presence of structurally abnormal ERCC6 (CSB) proteins, rather than the impaired tc-NER that results from antimorphic, hypomorphic and null mutations alike, and does not correlate with phenotype.
ACKNOWLEDGMENTS
We are grateful to our patients' families for supporting publication.




