Screening of CD96 and ASXL1 in 11 patients with Opitz C or Bohring–Opitz syndromes
Abstract
Opitz C trigonocephaly (or Opitz C syndrome, OTCS) and Bohring-Opitz syndrome (BOS or C-like syndrome) are two rare genetic disorders with phenotypic overlap. The genetic causes of these diseases are not understood. However, two genes have been associated with OTCS or BOS with dominantly inherited de novo mutations. Whereas CD96 has been related to OTCS (one case) and to BOS (one case), ASXL1 has been related to BOS only (several cases). In this study we analyze CD96 and ASXL1 in a group of 11 affected individuals, including 2 sibs, 10 of them were diagnosed with OTCS, and one had a BOS phenotype. Exome sequences were available on six patients with OTCS and three parent pairs. Thus, we could analyze the CD96 and ASXL1 sequences in these patients bioinformatically. Sanger sequencing of all exons of CD96 and ASXL1 was carried out in the remaining patients. Detailed scrutiny of the sequences and assessment of variants allowed us to exclude putative pathogenic and private mutations in all but one of the patients. In this patient (with BOS) we identified a de novo mutation in ASXL1 (c.2100dupT). By nature and location within the gene, this mutation resembles those previously described in other BOS patients and we conclude that it may be responsible for the condition. Our results indicate that in 10 of 11, the disease (OTCS or BOS) cannot be explained by small changes in CD96 or ASXL1. However, the cohort is too small to make generalizations about the genetic etiology of these diseases. © 2015 Wiley Periodicals, Inc.
INTRODUCTION
Opitz C syndrome (OTCS, also known as the C trigonocephaly syndrome, MIM #211750) is a rare disorder of trigonocephaly and associated hypertelorism, multiple buccal frenula, limb defects, wide alveolar ridges, visceral anomalies, redundant skin, hypotonia, psychomotor, and mental retardation. Less than 60 cases are known worldwide [Opitz et al., 2006].
Although the molecular basis of OTCS and its inheritance are unknown, this disorder was recently associated with mutations in the CD96 gene (Cluster of Differentiation 96, also called TACTILE for “T cell activation increased late expression,” MIM *606037) [Kaname et al., 2007]. In an OTCS individual, these authors discovered a balanced chromosomal translocation, t(3;18), that disrupted the CD96 gene at the 3q13.3 breakpoint. They also identified a missense mutation (c.839C > T, p.Thr280Met) in exon six of CD96 in one BOS patient. The CD96 gene encodes the CD96 antigen, a member of the immunoglobulin superfamily, which plays a role in cell adhesion [Fuchs et al., 2004]. Cells with mutated CD96 protein display loss of adhesion and growth activities in vitro [Kaname et al., 2007]. Moreover, the expression patterns of this gene, including bone and fetal brain are consistent with organs and tissues involved in the abnormalities of the C syndrome.
Recently, the role of CD96 in OTCS was questioned by Darlow et al. [2013]. These authors found a disruption of the CD96 gene in patients bearing a t(2;3) balanced translocation and who developed renal cell carcinoma without symptoms of OTCS. To reconcile both findings, it was proposed that the breakpoint region of chromosome 18 involved in the translocation found by Kaname et al. [2007] could bear the gene responsible for OTCS in that patient.
Regarding inheritance, some reports of affected sibs support autosomal recessive segregation [Antley et al., 1981; Opitz et al., 2006]. Likewise, normal karyotypes in most patients, unaffected parents with multi-affected offspring, an equal sex ratio of affected individuals, and consanguineous mating support autosomal recessive inheritance. However, many other patients are sporadic cases, which suggests the possibility of dominant inheritance or germ-line mosaicism [Sargent et al., 1985]. OTCS is highly variable, so that many symptoms that define OTCS overlap with Bohring–Opitz syndrome (BOS, MIM #605039). While some experts have defined these pathologies as two different conditions, others think that they are different presentations of the same disorder, ranging from a milder form (OTCS) to a more severe condition (BOS) [Bohring et al., 1999]. BOS is a rare malformation syndrome with only 30 cases described so far [Hastings et al., 2011]. It is characterized by exophthalmos, hypertelorism, severe intrauterine growth retardation, poor feeding, profound mental retardation, trigonocephaly, prominent metopic suture, nevus flammeus of the forehead, upslanting palpebral fissures, hirsutism, and flexion at the elbows, and wrists. The molecular basis of BOS and its inheritance are incompletely understood. It was associated with dominant mutations in the ASXL1 gene (Additional Sex Combs-Like 1; MIM *612990). ASXL1 gene maps to chromosome position 20q11.21 and it encodes the additional sex comb-like protein 1, which is a mammalian homolog of Drosophila asx. Recently, Hoischen et al. [2011] discovered that seven out of 13 BOS patients were heterozygotes for de novo mutations of this gene. Similar mutations in ASXL1 were found in two BOS patients by Magini et al. [2012]. Although BOS is considered an autosomal dominant pathology due to de novo mutations in ASXL1, the case of two sibs diagnosed with BOS [Greenhalgh et al., 2003], suggests that this malformation syndrome could also display an autosomal recessive pattern of inheritance (although gonadal mosaicism should also be considered in this case, consistent with AD inheritance).
In this study, CD96 and ASXL1 were analyzed in 11 patients with OTCS or BOS.
MATERIALS AND METHODS
Patients and Samples
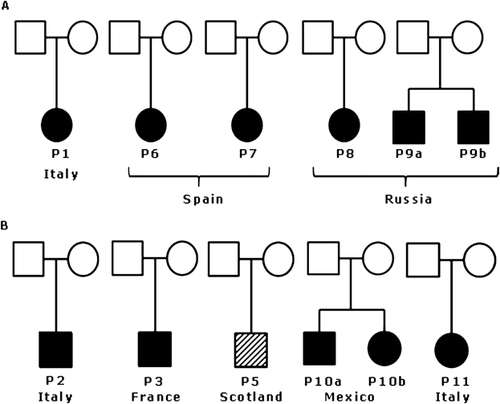
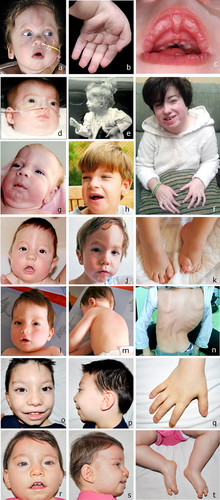
Most of the patients were diagnosed as OTCS or BOS by two of us (J.M.O., G.N.), and patients P9a and b, and P10 were diagnosed elsewhere (Fig. 1). Written informed consent was obtained from all patients’ parents. We have obtained the consent to publish pictures of patients P5, P6, P7, P9a, P9b, P10a, and P10b.
Genomic DNA was obtained from the patients and parents from peripheral blood using Wizard® Genomic DNA Purification Kit (Promega, Madison, WI). Quantity and quality of the gDNA was assessed using a Nanodrop ND-1000 Spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE).
Polymerase Chain Reaction (PCR) Amplification
Primers and PCR conditions for the amplification of the exons and flanking regions of CD96 and ASXL1 genes are summarized in Tables I and II, respectively. ASXL1 sequence ENST00000375687 and CD96 sequence ENSG00000153283 were taken as reference. Primers for DSC2 amplification were: DSC2_mut1_1-F: TTAGATCTTAATATTTGCCAGTG and DSC2_mut1_1-R: CATGCAGTGTTTGATTTTCTATT (Ta: 60°C); DSC2_mut2_1-F:TCTCCCCACGTGCATACATT and DSC2_mut2_1-R:CCATGTGACACAGCCCTTTT (Ta: 62°C).
| Primer name | Sequence 5′-3′ | Exon | Size (bp) | Ta (°C) | [MgCl2] (mM) |
|---|---|---|---|---|---|
| CD96-1F | CCAGGCGTGTGTGGTAGAGT | 1 | 277 | 62 | 1.5 |
| CD96-1R | CCTTGTGACTGCTGCTGAAA | ||||
| CD96-2F | TCTCATGCATCTCTATCTGAGTGA | 2 | 685 | 62 | 1.5 |
| CD96-2R | GACAGGAAGTGACCACAGCA | ||||
| CD96-3F | CCCTGAGGACAGATGAATCC | 3 | 365 | 62 | 1.5 |
| CD96-3R | ATTATGCCAGTGGTGGGAAG | ||||
| CD96-4F | AGGGGGTACCTGCCTTCTTA | 4 | 427 | 62 | 2 |
| CD96-4R | CATCCTCTGTTTCTGCCCAT | ||||
| CD96-5F | TCAGAAGAACAGAATGCTTCCA | 5 | 825 | 62 | 2.5 |
| CD96-5R | GGGGACTCTCTGAAGAATC | ||||
| CD96-6F | AGGTAGGATGACATTTGGTGC | 6 | 317 | 62 | 2 |
| CD96-6R | CTCTCAGAATGTTTGATGTGCA | ||||
| CD96-7F | GCCCATTTATAGTGCACGTTCT | 7 | 400 | 62 | 2 |
| CD96-7R | TCCAAGAAAAACATCATTCTGC | ||||
| CD96-8F | ACCATGGATCAATGACATGC | 8 | 568 | 62 | 2 |
| CD96-8R | TTTGACAAGTAGGACTCCAGTGA | ||||
| CD96-9F | TGATGAATAAATAAAGGAAAGTG | 9 | 654 | 58 | 2.5 |
| CD96-9R | CCAAAACAATGTTCTCAGTGG | ||||
| CD96-10F | GGCTGTTCACTAAGATTCTTTC | 10–11 | 942 | 62 | 2.5 |
| CD96-11R | TGAGACTACACCCATGGACAAA | ||||
| CD96-12F | TCCCACTTTGTTTGAGGACA | 12 | 397 | 62 | 2 |
| CD96-12R | GGAAACAAAATTCTTCTCAGCC | ||||
| CD96-13F | GGATCCCAGCCTTGCTATAA | 13 | 663 | 58 | 2.5 |
| CD96-13R | TGAGAAAGAAATCATTGGAACAAA | ||||
| CD96-14F | GAAAATTTCTGGTAGGGTACGTAAT | 14 | 585 | 62 | 2 |
| CD96-14R | GAGCAAGTCCGTGGACATG | ||||
| CD96-15F | TGCTGAAAGAACTGGGAGAG | 15 | 593 | 62 | 2 |
| CD96-15R | TGGTTTCAACTGTATGCCCA |
| Primer name | Sequence 5′-3′ | Exon | Size (bp) | Ta (°C) | [MgCl2] (mM) |
|---|---|---|---|---|---|
| ASXL1-1F | GAGCCGTGACCTCTGACC | 1 | 581 | 62 | 2 |
| ASXL1-1R | GAGGGCGAGCGGAAAAAGG | ||||
| ASXL1-2F | GGTCTCGCATACTATACCATG | 2 | 572 | 62 | 2.5 |
| ASXL1-2R | GGAAGAACAAGGAGATGGGG | ||||
| ASXL1-3F | TGTGGGGAGGGAAGATTGAG | 3 | 445 | 62 | 2.5 |
| ASXL1-3R | CCTTGGAACACACTGGATGC | ||||
| ASXL1-4F | AGTGTTAGTTATGGATTTCGGG | 4 | 475 | 62 | 2.5 |
| ASXL1-4R | CATCAGTGTCACCTGACTGC | ||||
| ASXL1-5F | GGTCAGTTAGAAAGGGTTAGG | 5–6 | 512 | 62 | 2.5 |
| ASXL1-6R | GCTCACTACAGGAGAAGGAG | ||||
| ASXL1-7F | ATAGTGTCGCCAGGGAATGC | 7–8 | 860 | 62 | 2.5 |
| ASXL1-8R | GACATCATCTTCTCACTAGGC | ||||
| ASXL1-9F | AGGAGATGCAACCCCAAGTG | 9–10 | 674 | 62 | 2.5 |
| ASXL1-10R | TAACCCTTTGAGAAGAGCGAG | ||||
| ASXL1-11F | CTCTGTCCCTATAAGAGCATG | 11 | 527 | 62 | 2.5 |
| ASXL1-11R | ATCACAGCCATCAGTTTCAGG | ||||
| ASXL1-12F | CCATTCATAGTGAAGCTAACAG | 12 | 875 | 62 | 2.5 |
| ASXL1-12R | GAGTTAAGGGCTCTATAATCTC | ||||
| ASXL1-13.1F | TGATTCTGTATGCCATGACCC | 13.1 | 797 | 62 | 2.5 |
| ASXL1-13.1R | TGCTCCTCATCATCACTTTCC | ||||
| ASXL1-13.2F | CTACTGTCCTCCCAAACCTC | 13.2 | 868 | 62 | 2.5 |
| ASXL1-13.2R | CCAGACGCATGTCACCATTC | ||||
| ASXL1-13.3F | TACAGCCTCTGACTTTGAAGG | 13.3 | 935 | 62 | 2.5 |
| ASXL1-13.3R | CAGCCACATTCCCAGAGCC | ||||
| ASXL1-13.4F | GAACACCTCGTTTCTCATCTC | 13.4 | 946 | 62 | 2.5 |
| ASXL1-13.4R | TGGGGGAAGGCAAGAGTGC |
The PCR reactions were carried out with 0.2 μM of each dNTP, 0.4 μM of each primer and 0.7 u of GoTaq Flexy (Promega) in the presence of 50 ng of gDNA. All the reactions consisted of an initial denaturalization step of 4 min at 95°C; 35 cycles of 30 sec at 95°C, 30 sec at annealing temperature (Ta), and 5 sec at 72°C, followed by a final extension of 4 min at 72°C. For exon1 of the ASXL1 gene, the GC-Rich PCR System (Roche, Basel, Switzerland) has been used. Briefly, 0.16 μM of each dNTP, 0.3 μM of each primer, 1 u of the GC-Rich Enzyme and 100 ng of gDNA. For the reactions, we performed an initial denaturalization step of 5 min at 95°C, 35 cycles of 30 sec at 95°C, 30 sec at 57°C, and 40 sec at 72°C, followed by a final extension of 5 min at 72°C.
The PCR products were purified with the MultiScreenTM Vacuum Manifold 96-well plate (Merck Millipore, Bellerica, MA) following the manufacturer's instructions and quantified using NanoDrop ND-1000 Spectrophotometer (Nanodrop Technologies Inc.).
Sanger Sequencing
The sequencing reactions were performed using the BigDye® Terminatior v3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA). The samples were processed at the Scientific and Technological Centers (CCiT) of the University of Barcelona.
Whole Exome Sequencing
Exome capture and sequencing was performed at CNAG, Barcelona, Spain. Briefly, the exons were captured by Agilent SureSelect Human All Exon kit (Santa Clara, CA) and sequenced by Illumina HiSeq2000 (San Diego, CA).
Base calling and quality control were performed using the Illumina RTA sequence analysis pipeline. Analysis of the primary data (FASTQ files) was done using the BIEŔs platform pipeline (CIBERER), which was as follows. Sequence reads were aligned to the human reference genome build GRCh37 (hg19) using the Burrows-Wheeler Aligner (BWA) [Li and Durbin, 2009]. Mapped reads were filtered (leaving only those mapping in unique genomic positions with enough quality), sorted and indexed with SAMtools [Li et al., 2009]. GATK [McKenna et al., 2010] was then used to realign the reads as well as for the base quality score recalibration. Once a satisfactory alignment was achieved, identification of single nucleotide variants and Indels was performed using GATK standard hard filtering parameters [DePristo et al., 2011]. For the final exome sequencing analysis report we used the VARIANT [Medina et al., 2012] annotation tool, which provides additional relevant variant information for the final process of candidate gene selection. In particular, minor allele frequency (MAF), obtained from dbSNP [Sherry et al., 2001] and 1,000 Genomes project [Abecasis et al., 2012], was provided to help on the selection of new variants not reported in healthy populations to date. SIFT [Kumar et al., 2009] and Polyphen [Adzhubei et al., 2010] damage scores were computed to predict the putative impact of the discovered variants over the protein structure and functionality. This information was completed with the evolutionary conservation obtained from PhastCons [Siepel et al., 2005]. Also disease related annotations, at both variant and gene levels were provided if available. Finally, GO terms for the affected genes were also retrieved. The successive application of quality control filters and the prioritization by the parameters accounting for potential functional impact led us to build up a list of candidate genes (and variants) ranked by its segregation with the cases and the putative potential impact. Such a prioritization list was further inspected to look for potential candidate genes and/or variants. Candidate variants were visualized by IGV software [Thorvaldsdottir et al., 2013].
RESULTS
Patients’ Phenotypes
The available clinical findings of the patients are summarized in Table III and pictures of most of them are shown in Figure 2. Patient P1 was previously reported (and pictures included) in Lalatta et al. [1990].
| P1 | P2 | P3 | P5 | P6 | P7 | P8 | P9a | P9b | P10a | P10b | P11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age at diagnosis (y) | 4 | 14 | NA | 2 | 2 | 2 | 4 | 0.5 | 0 | 1 | 0 | 6 |
| Consanguinity | No | No | NA | NA | No | No | No | No | No | No | No | No |
| MANIFESTATIONS CNS | ||||||||||||
| Developmental retardation | + | + | + | + | + | + | + | Very mild | No | + | + | + |
| Motor developmental delay | + | + | ||||||||||
| Seizures | + | + | + | + | ||||||||
| Brain MRI | (1) | (2) | (3) | (4) | Normal | (5) | ||||||
| HEAD | ||||||||||||
| Trigonocephaly | + | + | + | + | + | + | (6) | + | + | + | + | + |
| Facial anomalies | + | + | + | + | + | + | + | + | + | + | + | |
| Myopia | + | + | NA | |||||||||
| Strabismus | + | + | + | + | NA | |||||||
| Low-set and abnormally shaped ears | + | + | + | + | + | + | + | |||||
| Forehead hemangioma | + | + | ||||||||||
| Cutis marmorata/livedo reticularis | + | + | ||||||||||
| NECK | ||||||||||||
| Short, webbed neck | + | + | + | + | ||||||||
| THORAX | ||||||||||||
| Widely spaced nipples | + | + | ||||||||||
| Scoliosis | Mild | Severe | Severe | Severe | ||||||||
| LIMBS | ||||||||||||
| Short limbs | + | |||||||||||
| Flexion deformities | + | + | + | + | + | |||||||
| Brachydactyly | + | + | ||||||||||
| Single palmar crease | + | + | + | |||||||||
| Ulnar deviation of the fingers | + | + | + | |||||||||
| OTHER | ||||||||||||
| Generalized muscular atrophy | + | + | + | |||||||||
| Muscular hypotrophy | + | |||||||||||
| Hypotonia | + | + | + | + | + | + | + | + | + | |||
| Cryptorchidism | Bilateral | Bilateral | Left | Bilateral | ||||||||
| Horseshoe kidneys | + | |||||||||||
| Failure to thrive | + | + | + | |||||||||
- NA, not available. (1) Posterior thinning corpus callosum noted on MRI; (2) Mild enlargement of the supratentorial ventricular system, mainly in the frontal horns, and asymmetric lateral ventricles. Mild elevation of the choline in the spectroscopy. (3) A brain MRI showed moderate distension of the bodies and back horns of tricorns and of parieto-occipital subarachnoid spaces; (4) MRI CNS: Dimorphic ventriculomegaly with thin corpus callosum and white matter gliosis; (5) Brain MRI showed peritrigonal white matter thinner and mild ventricular enlargement; (6) mild brachycephaly, prominence of the metopic suture without definite trigonocephaly.
Search for Mutations Within the CD96 Coding Sequence
For the patients subjected to WES (Fig. 1A), the CD96 genomic region was analyzed from the BAM files using the IGV software. All exons were properly captured and sequenced (with an average coverage of 76.2 ± 13.7). Low quality changes detected were verified by Sanger sequencing and were discarded because they were false positives. For the remaining five patients, the 15 coding exons and their flanking regions were Sanger sequenced. No mutation was found in any of the 11 patients included in this study.
Search for Mutations Within the ASXL1 Coding Sequence
The genomic region of ASXL1 was analyzed in silico by IGV in the six patients included in the exome project. The average coverage for the ASXL1 gene in these samples was 34.6 ± 6.7. Exons 1, 7, 8, and the 5′ fragment of exon 13 (13.1) were poorly captured and, thus, were sequenced by the Sanger method in all the patients. In addition, the 13 exons and flanking regions were Sanger-sequenced in the patients not included in the exome project.
In patient P5's exon 13 we found a heterozygous 1 bp duplication (c.2100dupT, p.Pro701Serfs*16) absent in his parents. It was thus, a de novo mutation. No other mutation was found in the remaining patients.
Analysis of Genomic Region 18q12.1
For the patients subjected to WES, we analyzed by IGV a 3.22 Mb region between chr18:28495126 and chr18:31717104 that includes the desmocollin (DSC) and desmoglein (DSG) gene clusters. In particular, the exons of 20 genes within the region, including ASXL3, were analyzed (Table IV). For this region, we have an average coverage of 70.3 ± 35.7). The only changes found were two missense mutations in the DSC2 gene (p.Glu46Lys and p.Glu102Lys) in a compound heterozygous state in patient P1. Mutation p.Glu46Lys (rs180908546; MAF: 0.0005 in 1000 Genomes and MAF: 0.00007 in ESP6500) was inherited from the patient's father while p.Glu102Lys (rs144799937; MAF: 0.00076 in ESP6500) was inherited from her mother.
| DSC2 | DSG2 | TRAPPC8 | WBP11P |
| DSC1 | TTR | RNF125 | KLHL14 |
| DSG1 | B4GALT6 | RNF138 | C18orf34 |
| DSG4 | SLC25A52 | MEP1B | ASXL3 |
| DSG3 | MCART2 | FAM59A | NOL4 |
DISCUSSION
Little is known of the molecular bases of the OTCS and BOS syndromes. To date, only one gene, CD96, has been associated with OTCS by Kaname et al. [2007], who reported an OTCS patient bearing a balanced chromosomal translocation, t(3;18)(q13.13;q12.1) that disrupted CD96 at 3q13. The precise breakpoint was within CD96 exon five and led to a premature termination of the protein. In that patient, CD96 mRNA expression was reduced to 50%. Moreover, in the mutation analysis of nine karyotypically normal patients diagnosed with OTCS or BOS, Kaname et al. [2007] also identified a missense mutation (c.839C > T, p.Thr280Met) in one BOS patient, not found in controls. In our study, we did not find any CD96 mutation in the 11 patients included in the present study.
The relationship between CD96 and Opitz C syndrome was questioned recently as this gene was found disrupted in patients with renal cell carcinoma without any resemblance to Opitz C patients [Darlow et al., 2013]. Alternatively, it was proposed that in Kaname's patient, the breakpoint region on chromosome 18 (18q12.1) may be the one bearing the OTCS causative mutation. This region in now known to comprise the desmocollin and desmoglein gene clusters, responsible for tight junction formation in epithelia. Additionally, it includes the ASXL3 gene (a paralog of ASXL1), which was recently associated with a syndrome with phenotypical overlap with BOS [Bainbridge et al., 2013]. It should be very interesting to sequence this gene in a sample from the patient with the t(3;18) translocation described by Kaname et al. [2007].
In our study, we scrutinized the exon sequences within 3.22 Mb of chromosome region 18q12.1 of the six patients subjected to WES. In patient P1 (OTCS) we found two mutations in the DSC2 gene (p.Glu46Lys and p.Glu102Lys). Both mutations are present in the general population albeit with a low frequency (MAFs <0.001) and both are classified as benign in Polyphen2 (with scores of 0.006 and 0.016, respectively). The p.Glu102Lys mutation was reported previously in a heterozygous state in a patient with arrhythmogenic right ventricular cardiomyopathy (ARVC) [Anastasakis et al., 2012]. The clinical manifestations of patient P1 were previously reported by Lalatta et al. [1990] (as patient 3; C.L.), and at age 4 years, this patient did not present any kind of cardiomyopathy. However, ARVC is usually manifest in late adulthood. Interestingly, Andreasen et al. [2013] have pointed out that a large number of the mutations reported as associated with a cardiomyopathy are not, in fact, the monogenic cause of the disease. Many of these changes (14–18%), including the p.Glu102Lys mutation, are found in public databases such as the NHLBI Exome Sequencing Project (ESP). According to the observed frequency of these mutations in ESP, the expected frequency of ARVC should be 1:5, a prevalence far higher than that observed in the general population, which is estimated as 1:5000. Also, when compared to the ones not present in the public databases, the mutations found in ESP differ significantly in the number of nonsense mutations (lower) and in the PolyPhen2 prediction (often classified as “benign”). All things considered, we understand that the DSC2 mutations found in patient P1 are not responsible of her OTCS phenotype. No other putative pathogenic mutation was found in any of the remaining exons of the genes in the 18q12.1 region in the remaining patients included in the exome project. This includes ASXL3, a very interesting candidate. However, the exome analysis does not allow for the inspection of regulatory sequences located outside the exons.
In our study we have assessed the ASXL1 gene, both in OTCS and BOS patients, and we have found a loss-of-function mutation in one patient with a clear diagnosis of BOS (P5). This mutation (c.2100dupT, p.Pro701Serfs*16) consists of an insertion of a T at position 2100 of the cDNA, leading to a change in the amino acid at residue 701 and a premature truncation of the protein. This change was found in patient P5 but was absent in his parents, being a de novo change. This mutation is located in exon 13, as all but one (p.Arg404X) of the ASXL1 mutations previously associated with BOS [Hoischen et al., 2011; Magini et al., 2012]. And, as all of them, it leads to a premature end of the protein. All these facts suggest that this mutation is the cause of the BOS phenotype in the P5 patient.
In summary, we have not found any mutation in the CD96 gene or in the 18q12.1 region that could explain the OTCS or BOS phenotypes of our patients. In contrast, we have found a de novo mutation in the ASXL1 gene in the BOS patient, adding a novel mutation to those described for BOS and found no ASXL1 mutation in OTCS patients. Phenotypically, it is hard to define salient features allowing a clear distinction between these two conditions, except perhaps for the severity, which tends to be more marked in the BOS. The genes might provide a better distinction. For example, ASXL1 mutations were found only in BOS. But half of the BOS patients remain undiagnosed at the molecular level. However, it may happen that as soon as there is a new gene for OTCS, a patient with mutation in this gene will be considered BOS by some clinicians. Further analysis of the exome sequencing, in a hypothesis-free approach, currently underway, should help shed light on the molecular bases of OCTS.
ACKNOWLEDGMENTS
The authors thank the patients and their families for their wholehearted collaboration and Dr. Karen Naismith, Dr. María del Carmen Esmer, Dr. Francina Munell, and Dr. María Amparo Alonso for sharing clinical information of specific patients. They are also grateful for the technical assistance of Ms. M. Cozar. Funding was from Associació Síndrome Opitz C, Terrassa, Spain; Spanish Ministerio de Ciencia e Innovación (SAF2011-25431; SAF2014-56562-R; FECYT, crowdfunding PRECIPITA); Catalan Government (2009SGR971, 2014SGR932), and from CIBERER (U720).