MED23-associated intellectual disability in a non-consanguineous family
Abstract
Intellectual disability (ID) is a heterogeneous condition arising from a variety of environmental and genetic factors. Among these causes are defects in transcriptional regulators. Herein, we report on two brothers in a nonconsanguineous family with novel compound heterozygous, disease-segregating mutations (NM_015979.3: [3656A > G];[4006C > T], NP_057063.2: [H1219R];[R1336X]) in MED23. This gene encodes a subunit of the Mediator complex that modulates the expression of RNA polymerase II-dependent genes. These brothers, who had profound ID, spasticity, congenital heart disease, brain abnormalities, and atypical electroencephalography, represent the first case of MED23-associated ID in a non-consanguineous family. They also expand upon the clinical features previously reported for mutations in this gene. © 2015 Wiley Periodicals, Inc.
INTRODUCTION
Intellectual disability (ID) is a significant limitation in cognitive functioning and a lack of adaptive skills necessary for daily living [APA, 2013]. As a neurodevelopmental disorder with a substantial lifelong burden, ID adversely influences the affected individual, the family and/or caretakers, and the educational system. The diagnosis of ID involves: 1) deficits in intellectual functioning (IQ <70); 2) limitations in adaptive skills, such as communication, social participation, and independent living; and 3) onset of symptoms before 18 years of age [APA, 2013]. Depending on the level of impairment in conceptual, social and practical domains, ID can be classified as mild, moderate, severe, or profound. In addition, ID is categorized as syndromic or nonsyndromic based on the presence or absence of co-morbid features [Kaufman et al., 2010a].
Recent reports have linked ID and other related neurological disorders to mutations in more than 450 different genes, many of which encode proteins essential for transcriptional regulation, such as components of the Mediator complex [Van Bokhoven, 2011]. This complex is a multi-subunit assembly that relays signals from upstream enhancer elements to downstream RNA polymerase II machinery. Mutations in subunits of the Mediator complex, including MED12, MED17, and MED25, have been associated with cognitive impairment, brain abnormalities and/or neuromuscular deficits [Risheg et al., 2007; Schwartz et al., 2007; Leal et al., 2009; Kaufman et al., 2010b]. A large consanguineous Algerian family showed cosegregation of nonsyndromic autosomal recessive ID with a homozygous mutation in MED23, the gene encoding a subunit within the tail region of the Mediator complex [Hashimoto et al., 2011]. This mutation disrupted the ability of MED23 to interact with enhancer-bound transcription factors, TCF4 and ELK1, resulting in the dysregulation of immediate early genes (JUN and FOS) necessary for brain development and neuroplasticity.
We present two male siblings with profound ID and novel disease-segregating mutations in MED23. These brothers, who are the first MED23-deficient patients reported from a nonconsanguineous family, expand the phenotype associated with mutations in this gene to include profound ID, spasticity, congenital heart disease, brain abnormalities, and atypical electroencephalography (EEG).
MATERIALS AND METHODS
Patients
The propositi were admitted to the National Institutes of Health Clinical Center (NIH-CC) and enrolled in protocol 76-HG-0238, “Diagnosis and Treatment of Patients with Inborn Errors of Metabolism or Other Genetic Disorders,” approved by the National Human Genome Research Institute (NHGRI) Institutional Review Board (IRB). After parents gave informed consent, the children were investigated at two different time points, i.e., ages 8 and 11 years for Patient 1 and ages 23 months and 5 years for Patient 2.
Exome Analysis
Genomic DNA was extracted from peripheral whole blood of the two affected individuals, their unaffected parents, and two unaffected siblings using the Gentra Puregene Blood Kit (Qiagen, Valencia, CA). The DNA of all family members was subjected to an integrated set of genomic analyses including high-density single-nucleotide polymorphism (SNP) arrays and whole exome sequencing (WES). WES was performed on the nuclear family using the Illumina HiSeq2000 platform and the TrueSeq capture kit (Illumina, San Diego, CA). Sample library preparation, sequencing, and analysis were performed using the standard NIH Intramural Sequencing Center (NISC) pipeline [NISC Comparative Sequencing Program, 2014]. Sequence data was aligned to human reference genome (hg19) using Novoalign (Novocraft Technologies, Selangor, Malaysia). To test for copy number variants and to form segregation BED files for exome analysis, Omni Express 12 (hg18) SNP arrays were run on genomic DNA from all family members as described [Markello et al., 2012] and converted to hg19. Variants listed in the Variant Call Files (VCFs) were filtered based on rarity, Mendelian segregation, and predicted deleteriousness. Allele frequencies were required to be <0.06 for the UDP patient cohort and <0.02 for the Exome Sequencing Project v.0.0.20 African Ancestry population, Exome Sequencing Project v.0.0.20 European Ancestry population, and dbSNP build 137. Also, homozygous and de novo variants were required to occur in < 2 individuals in the UDP cohort (excluding the affected brothers). Variants needed to segregate with autosomal recessive, X-linked recessive, or de novo dominant modes of inheritance and be annotated as nonsynonymous, frame shift, premature stop, loss of start codon, loss of stop codon, near splice (within 20 base pairs of a canonical splice site), or splicing mutations. Variants that passed filtration were ranked using Exomiser 2.0 (http://www.sanger.ac.uk/resources/databases/exomiser/q uery/exomiser2); the shared Human Phenotype Ontology terms of the patients were used as the phenotype input for Exomiser. Using the Integrative Genome Viewer (https://www.broadinstitute.org/igv/home), we assessed the quality of alignment and genotype call of variants.
Sanger DNA sequencing was used to validate the variants identified. Using the primers listed in Supplementary Table I (in supporting information online), PCR amplification was performed using Qiagen HotStarTaq master mix (Qiagen, Valencia, CA). The following conditions were used for amplification: 1 cycle of 95°C for 5 min, followed by 39 cycles of 95°C for 30 sec, 55°C for 30 sec, 72°C for 30 sec, and a final extension at 72°C for 5 min. Unincorporated primers and nucleotides were removed using ExoSAP-IT reagent (USB, Cleveland, OH). Sanger sequencing of the PCR products was performed by Macrogen (Rockville, MD). The sequences were aligned and analyzed using Sequencher v.5.0.1 (Gene Codes, Ann Arbor, MI). CLIA certification was performed by Sanger DNA sequencing (Gene Dx, Rockville, MD).
Gene Expression Analysis
Skin fibroblasts were obtained from both patients under NHGRI IRB- approved protocols. Control skin fibroblasts were procured from American Type Culture Collection (Manassas, VA) or Coriell (GM08398, Camden, NJ). Patient and control fibroblasts were grown in high- glucose DMEM (Life Technologies, Carlsbad, CA) supplemented with 10% fetal bovine serum and 1% Antibiotic-Antimycotic (Life Technologies, Carlsbad, CA). Cultured fibroblasts were incubated in a humidy controlled environment at 37°C, with 95% O2 and 5% CO2, fed fresh media every 3 days, and were used before passage 10.
Total RNA was isolated from each cell line according to the instructions of the RNeasy Mini Kit (Qiagen, Valencia, CA). Purified RNA was reverse transcribed using First- strand cDNA Synthesis (Origene, Rockville, MD). Real- time quantitative PCR was carried out on the ABI PRISM 7000 Sequence Detection System using TaqMan Universal PCR Master Mix and TaqMan Gene Expression Assays for human MED23 (Hs00606608_m1), for human FOS (Hs04194186_s1), for human JUN (Hs01103582_s1), and GAPDH (Hs02758991_g1) (Life Technologies, Grand Island, NY). Expression levels of each transcript were normalized to GAPDH.
Allele frequencies were quantified using the RainDrop Digital PCR System (RainDance Technologies, Inc., Billerica, MA). Custom TaqMan SNP Genotyping Assays were designed for both mutations (LifeTechnologies, Grand Island, NY). Digital PCR was performed on patient and control cDNA according to the manufacturer's instructions. Briefly, the 40 μl reaction mixture (50 ng cDNA, 1X TaqMan SNP Genotyping Assay, 1X TaqMan Genotyping Master Mix, and 1X Droplet Stabilizer) was emulsified into uniform droplets using the RainDrop Source machine. PCR amplification was then performed in a BioRad MyCycler thermal cycler (BioRad Laboratories, Hercules, CA) using a 0.5°C per sec ramp speed and the following cycling parameters: 10 min at 95°C, then 40 cycles of 95°C for 15 sec and 60°C for 1 min, followed by an additional 10 min at 95°C. The resulting fluorescence of three million droplets per reaction was measured on the RainDrop Sense machine and allele frequencies were calculated using RainDance Analyst software (RainDance Technologies, Inc., Billerica, MA).
Western Blot Analysis
Nuclear proteins were isolated from patient and control skin fibroblasts according to the instructions of the Qproteome Cell Compartment Kit (Qiagen, Valencia, CA). Proteins were then resolved by SDS-PAGE and transferred to a nitrocellulose membrane. The blot was incubated with antibodies against MED23 (BD Pharmingen, San Jose, CA) and histone H2B (EMD Millipore, Billerica, MA) and visualized using the Odyssey CLx infrared imaging system (LI-COR Biosciences, Lincoln, NE).
CLINICAL REPORT
Patient 1
Patient 1 was an 11 -year-old Caucasian male who presented to the Undiagnosed Diseases Program (UDP) at the National Institutes of Health (NIH) with profound developmental delay, spasticity, and static encephalopathy. He was the eldest of four brothers in a nonconsanguineous family of maternal Hungarian and paternal Czechoslovakian and Polish ethnicity. Notable features, such as birth measurements and abnormal behavioral findings from infancy are described in Table I. He had a small ventricular septal defect (VSD) and patent foramen ovale that closed spontaneously.
| Feature | Patient 1 | Patient 2 | Hashimoto et al. [2011] |
|---|---|---|---|
| Birth data | |||
| Gestation | 40 weeks | 36 6/7 weeks (twin) | NR |
| BW (Weight-for-age centile)* | 3 kg (50%) | 2 kg (5%) | NR |
| BL (Length- for-age centile)* | 55 cm (95%) | 50 cm (50%) | NR |
| OFC (Head circumference-for-age centile)** | 36 cm (50%) | 33 cm (10%) | NR |
| Apgar scores | 8/9 | 5/4 | NR |
| Behavioral findings | |||
| Infant irritability | + | + | NR |
| Difficulty feeding in infancy | + | + | NR |
| Sleep dysregulation in infancy | + | + | NR |
| Childhood screaming spells | + | + | NR |
| Cardiac | |||
| CHD | VSD | ASD | NR |
| Current growth measurements | |||
| Height-for- age centile* | 3–10% | 25–50% | normal |
| Weight-for- age centile* | 10–25% | 25–50% | normal |
| OFC (Head circumference-for-age centile)** | 54 cm (75%) | 49 cm (5%) | normal |
| Neurological findings | |||
| Cognitive impairment | profound ID | profound ID | mild-to-moderate ID |
| Degree of assistance in ADL | total | total | limited |
| Static encephalopathy | + | + | NR |
| Expressive language | – | – | + |
| Spasticity | + | + | NR |
| Axial hypotonia | + | + | NR |
| Dystonia | + | – | NR |
| Choreoathetoid movements | – | + | NR |
| Abnormal electroencephalography | + | + | − |
| Brain imaging | |||
| Pontine hypoplasia | + | + | – |
| Thin corpus callosum | – | + | – |
| Temporal lobe hypomyelination | – | + | – |
- NR, Not Reported; VSD, Ventricular Septal Defect; ASD, Atrial Septal Defect; ADL, Activities of Daily Living.
- * Length and Weight-for-age centiles were calculated using individual growth charts provided by the Centers for Disease Control and Prevention [Kuczmarski et al., 2002].
- ** Current Head Circumference-for-age centiles were calculated using the Handbook of Normal Physical Measurements [Hall et al., 1989].
From early infancy, he exhibited globally delayed development without regression. He rolled over at age 2 years, sat by age 3 years, scooted at age 5 years, and walked with the assistance of a walker and ankle foot orthoses by age 6 years. On the Vineland Adaptive Behavioral Scales [Sparrow et al., 2005] at age 11 years, he scored below the first centile for chronological age in communication, daily living skills, socialization, and adaptive behavior composite. Concurrent testing with the Mullen Scales of Early Learning [Mullen, 1995] placed him at the age-equivalent of 14 months for visual reception, 8 months for fine motor, 10 months for receptive language, and 4 months for expressive language.
Muscle spasticity and abnormal movements began in infancy and led to a clinical diagnosis of cerebral palsy by age 1 year. Remarkable neurologic features at age 11 years are described in Table I. His markedly delayed fine motor skills impaired self-feeding, although he demonstrated willingness and ability to learn. After thorough cognitive and motor phenotyping, he was given the diagnosis of profound intellectual disability with co- morbid motor impairments [APA, 2013].
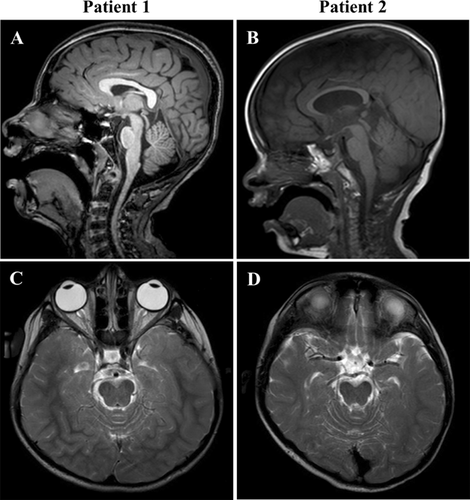
Abnormal brain imaging and EEG studies at age 11 years are summarized in Table I and Figure 1. The EEG was indicative of mild cerebral dysfunction and predisposition to partial or generalized seizures due to unprovoked and photic-provoked epileptiform abnormalities.
Patient 2
Patient 2 was the 5 -year-old brother of Patient 1. He presented to the UDP at 22 months with profound developmental delay, spasticity, and poor visual behavior. Birth information and behavioral findings from infancy are listed in Table I. He had a moderate secundum atrial septal defect (ASD) with right ventricular dilation and volume overload which resolved spontaneously. Eye examinations in the first year of life showed brief fixation, inability to track in any direction, and intermittent right exotropia.
Similar to his older brother, Patient 2 had global developmental delay that was exacerbated by visual cortical impairment. On the Vineland Adaptive Behavior Scales [Sparrow et al., 2005], he scored below the first percentile in communication, daily living skills, socialization, and adaptive behavior composite at age 5 years. On the Mullens Scales of Early Learning [Mullen, 1995], he obtained an age-equivalent of 3 months for visual reception, 2 months for fine motor, 4 months for receptive language, and 7 months for expressive language.
He also had muscle spasticity and abnormal movements leading to a clinical diagnosis of cerebral palsy by age 1 year. Significant neurologic findings are listed in Table I. At 22 months, he maintained a predominantly flexor position, but could lift his head and shoulders when prone, pull himself forward slightly using his elbows and roll to each side. By age 5 years, he maintained a sitting position with minimal support. After thorough phenotyping, he was diagnosed with profound intellectual disability with co-morbid motor and visual impairments [APA, 2013].
Imaging and EEG anomalies at age 5 years are summarized in Table I and Figure 1. His EEG indicated moderate-to-severe cerebral dysfunction with a disorganized slow background and predisposition to partial or generalized seizures due to bilateral frontal predominant epileptiform abnormalities.
RESULTS
To identify putative genomic causes for their disease, the propositi underwent array CGH and WES. The array CGH did not identify any pathological deletions or duplications. WES identified variants in 12 genes segregating with disease and meeting frequency and predicted deleteriousness requirements (Supplementary Table II). Ten mutations did not validate by Sanger sequencing (Supplementary Table I) leaving the compound heterozygous mutations for CFCTL and MED23. We considered the mutations in CFCTL an unlikely cause of disease because it had no known disease association, and because one mutation was intronic and the other was a polymorphism (frequency: 1–3%, rs146560816). In contrast, MED23 had been associated with disease and had compound heterozygous mutations (NM_015979.3: c. [3656A>G];[4006C>T]) predicted to encode a deleterious missense mutation (NP_057063.2: p.[H1219R],) and a premature stop codon (p.[R1336X]) (Fig. 2).
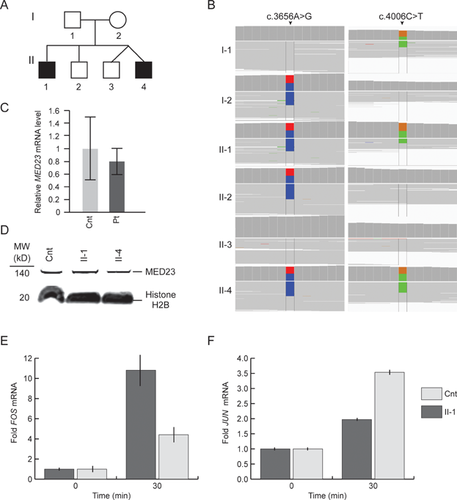
The father was heterozygous for c.4006C>T (p.R1336X), and the mother was heterozygous for c.3656A>G (p.H1219R). Real-time quantitative PCR of RNA isolated from patient and control skin fibroblasts detected no marked reduction in steady state levels of (mRNA) MED23 (Fig. 2). Allele-specific digital PCR confirmed a 1:1 ratio of steady state mRNA from the two mutant MED23 alleles (data not shown), further excluding nonsense-mediated mRNA decay. Immunoblot analyses of nuclear lysates revealed no alteration in MED23 steady state protein levels (Fig. 2); the resolution of the immunoblot was insufficient to detect whether a stable truncated MED23 protein (150 kDa versus 147 kDa) was produced from the c.4006C>T (p.R1336X) allele.
Confirming the functional significance of the identified mutations, cultured skin fibroblasts from Patient 1 had a similar alteration of the immediate early gene response to serum mitogens as the cultured fibroblasts of the previously reported patients [Hashimoto et al., 2011]. Using qRT-PCR, we observed down regulation of JUN expression and up regulation of FOS expression (Fig. 2 E and F).
DISCUSSION
We report the first nonconsanguineous family in which ID cosegregated with compound heterozygous mutations in MED23. The mutations (p.H1219R and p.R1336X) did not alter steady-state protein levels in cultured skin fibroblasts suggesting that both mutant proteins are incorporated into the Mediator complex. Similarly, the homozygous p.R617Q MED23 mutation, reported in five affected individuals of a consanguineous Algerian family, did not alter the stability of MED23 or the Mediator complex.
Although the propositi overlap phenotypically with the previously reported cases, there are some important differences. The two Algerian probands, age 39 and 41, had mild-to- moderate ID affecting only their ability to read, write, and look after their financial affairs; they had no pathological physical, brain imaging, or EEG findings [Hashimoto et al., 2011]. In contrast, our patients were completely dependent for all activities of daily living and had spasticity, cardiac anomalies, brain malformations, abnormal EEGs, and visual impairment (patient 2).
In addition to neurodevelopment, MED23 has a role in cardiovascular development. In zebrafish, Med23 knockdown causes shortened body length, a ventrally curved tail, heart edema, and bradycardia [Yin et al., 2012]. Knockout of (MOUSE) Med23 was embryonic lethal between E 9.5 and 10.5 due to systemic circulatory failure [Balamotis et al., 2009]. Other subunits of the Mediator complex have also been shown to affect cardiovascular development in murine model systems. (MOUSE) Med1 null embryos develop cardiac malformations, particularly ventricular hypoplasia [Landles et al., 2003], and (MOUSE)Med12 hypomorphic mutants fail to develop beyond E 10 due to severe defects in neural tube closure, axis elongation, and heart formation [Rocha et al., 2010]. Consequently, the mutations in (HUMAN) MED23 might explain the congenital heart defects of the propositi.
In summary, the propositi expand the phenotype observed with mutations in MED23 to include severe ID and spasticity, cardiac anomalies, brain malformations, and visual impairment (Patient 2 only). In addition, these patients illustrate the power of exome sequencing to identify the genetic basis of disorders without established pathognomonic features.
ACKNOWLEDGMENTS
The authors wish to thank the patients, their family and the treating physicians for their cooperation, encouragement and interest in the development of this manuscript.