Delineation of the KIAA2022 mutation phenotype: Two patients with X-linked intellectual disability and distinctive features
Abstract
Next-generation sequencing has enabled the screening for a causative mutation in X-linked intellectual disability (XLID). We identified KIAA2022 mutations in two unrelated male patients by targeted sequencing. We selected 13 Japanese male patients with severe intellectual disability (ID), including four sibling patients and nine sporadic patients. Two of thirteen had a KIAA2022 mutation. Patient 1 was a 3-year-old boy. He had severe ID with autistic behavior and hypotonia. Patient 2 was a 5-year-old boy. He also had severe ID with autistic behavior, hypotonia, central hypothyroidism, and steroid-dependent nephrotic syndrome. Both patients revealed consistent distinctive features, including upswept hair, narrow forehead, downslanting eyebrows, wide palpebral fissures, long nose, hypoplastic alae nasi, open mouth, and large ears. De novo KIAA2022 mutations (p.Q705X in Patient 1, p.R322X in Patient 2) were detected by targeted sequencing and confirmed by Sanger sequencing. KIAA2022 mutations and alterations have been reported in only four families with nonsyndromic ID and epilepsy. KIAA2022 is highly expressed in the fetal and adult brain and plays a crucial role in neuronal development. These additional patients support the evidence that KIAA2022 is a causative gene for XLID. © 2015 Wiley Periodicals, Inc.
INTRODUCTION
X-linked ID (XLID) accounts for 5–10% of intellectual disability (ID) in males. Mutations causing XLID have been reported in over 100 genes [Tarpey et al., 2009; Lubs et al., 2012]. High-throughput sequencing has enabled the screening for a causative mutation in XLID using targeted panel, X-exome, and whole exome sequencing methods [de Ligt et al., 2012; Nava et al., 2012]. Cantagrel et al. [2004] previously identified an inactivating disruption of the X-linked KIAA2022 and P2RY8 in two male patients with severe ID [Cantagrel et al., 2004]. Van Maldergem et al. [2013] reported three additional families with moderate to severe ID with truncated KIAA2022 mutations and 70-kb microduplication encompassing KIAA2022 exon 1 [Van Maldergem et al., 2013]. Here, we report two unrelated male patients with nonsyndromic ID and novel KIAA2022 nonsense mutations detected by targeted sequencing.
CLINICAL REPORTS
We selected 13 Japanese male patients, including four sets of siblings (three male siblings and one female sibling) and nine sporadic patients with severe ID and slightly dysmorphic features inconsistent with established syndromes, after exclusion of chromosomal anomalies by array comparative genomic hybridization (CGH). Two of thirteen patients had KIAA2022 mutation.
Patient 1
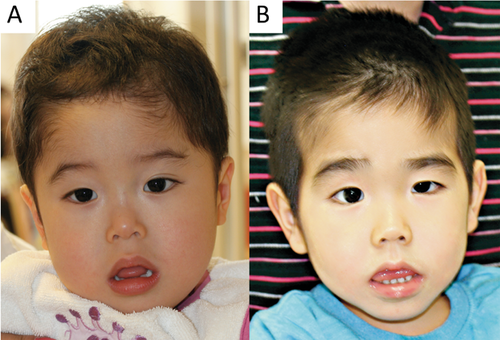
Patient 1 was a 3-year-old boy. He was the first and only child of unrelated healthy parents with no family history of ID. He was delivered at 33 weeks gestation owing to premature rupture of membrane after an uneventful pregnancy. His birth weight was 1762 g [− 0.5 standard deviation (SD)], length was 40.5 cm (− 1.3 SD), and OFC was 30.0 cm (0.4 SD). He was admitted to the neonatal intensive care unit and was supplied oxygen because of respiratory distress. Hypotonia was noted at birth, associated with poor feeding and poor weight gain. After discharge without tube feeding, he had failure-ro-thrive due to gastroesophageal reflux with persistent vomiting more than twice daily. The patient started controlling his head at 6 months, rolling over at 9 months, and sitting with support at 21 months. He was referred at 1 year and 9 months for assessment of hypotonia and developmental delay. His height was 73.6 cm (− 3.3 SD), weight 9.8 kg (− 1.2 SD), and OFC45.6 cm (− 1.7 SD). He had postnatal growth retardation (Suppl. Fig. S1A,B). He was able to sit alone at age three years; however, he did not speak comprehensively. He had autistic behavior with repetitive hand flapping movement and tongue protrusion. The patient showed dysmorphic features, including upswept hair, narrow forehead, downslanting eyebrows, long nose, hypoplastic alae nasi, thin lips, open mouth, and large ears (Fig. 1A). Brain magnetic resonance imaging (MRI) revealed normal structure for his age.
Patient 2
Patient 2 was a 5-year-old boy. He was the first child of unrelated healthy parents with no family history of ID. He had a young brother with normal development. He was delivered at 38 weeks and 6 days gestation after an uneventful pregnancy. His birth weight was 2948 g (− 0.3 SD), length 49.3 cm (0.1 SD), and OFC 31.0 cm (1.5 SD). He was referred at age 3 months for assessment of hypotonia. He started sitting at age 14 months, standing with support at 33 months, and walking alone at four years. The patient presented with steroid-dependent nephrotic syndrome at 3 years, with four relapses in 9 months. He then received cyclosporine, and remission was sustained. The patient developed central hypothyroidism [thyroid-stimulating hormone (TSH) level: 9.93 µIU/ml, free triiodothyronine level: 1.17 pg/ml, free thyroxine level: 0.52 pg/ml] requiring thyroid hormone replacement at age 4 years. A thyroid-releasing hormone (TRH) tolerance test revealed a TSH peak of 27.42 µU/L at 30 min. A pituitary function test (insulin, gonadotropin releasing hormone, TRH tolerance tests, and arginine tolerance test) revealed normal patterns other than thyroid hormone. At age 5 years, his height was 86.7 cm (− 5.0 SD), weight 10.1 kg (− 3.0 SD), and OFC 46.5 cm (− 2.9 SD). He had postnatal growth retardation (Suppl. Fig. S1C,D). He had severe developmental delay (development quotient of 20 at age 5 years) and did not speak at age 5 years. He had dysmorphic features, including upswept hair, narrow forehead, bushy and downslanting eyebrows, long nose, hypoplastic alae nasi, thin lips, open mouth, and large ears (Fig. 1B). The patient's karyotype was 46, XY. Electromyogram, electroencephalogram, and head MRI revealed normal findings. Blood amino acid and urine organic acid tests showed normal results.
MATERIALS AND METHODS
Written informed consent was obtained from the patients' parents in accordance with the Kanagawa Children's Medical Center Review Board and Ethics Committee.
Total genomic DNA was obtained from lymphocytes using standard techniques. Targeted panel was designed for targeted next-generation sequencing for 68 XLID genes and 101 autistic spectrum disorder (ASDs) genes, covering all coding regions and untranslated regions (in total, 1,164,259 bp). Candidate genes were selected from a literature review of XLID and ASDs genes associated with nonsyndromic ID [Vissers et al., 2010; Najmabadi et al., 2011; Lubs et al., 2012]. The list of genes is described in the supplemental data (Suppl. Table SI). DNA libraries were enriched for sequences using HaloPlex (Agilent Technologies Inc., Santa Clara, CA). Patient samples were sequenced by MiSeq (IlluminaInc., San Diego, CA) by 150-bp pair-end reads. Data were analyzed by Burrows–Wheeler alignment tool version 6 and Genome Analysis Toolkit pipeline (Broad Institute, Cambridge, MA) and visualized in the Integrative Genomics Viewer (IGV). Calling copy-number variation (CNV) was based on log–ratio analysis and the z-score of read depth on each exon. Mutations identified by targeted sequencing were confirmed by Sanger sequencing and demonstrated appropriate segregation with phenotype in unaffected mothers of each XLID case.
Array CGH was performed using the Agilent SurePrint G3 Human CGH Microarray Kit 8 × 60k (Agilent Technologies) according to manufacturer's instructions. The results were analyzed using Agilent Genomic Workbench software (Agilent Technologies).
RESULTS
The mean depth of coverage over all samples was 281× per base, and bases covered by at least five reads were 96.7% and those covered by at least 20 reads were 90.1% of coding regions. Targeted sequencing identified two novel mutations of KIAA2022 in two unrelated patients. Patient 1 had c.2113C>T/p.Q705X (NM_001008537) in the coding region of exon 3, and his mother did not have the mutation. Patient 2 had c.964C>T/p.R322X (NM_001008537) in the coding region of exon 3, and his mother did not have the mutation (Suppl. Fig. S2). Neither mutation was present in the human genetic variation database (Japanese genetic variation consortium, a reference database of genetic variations in Japanese population: 1,208 individuals http://www.genome.med.kyoto-u.ac.jp/SnpDB), the 1000 genomes project, and NHLBI grant opportunity exome sequencing project (ESP).
Array CGH revealed no significant copy number variants in either patient. In Patient 1, array CGH revealed an 886-kb duplication at 4q28.3 from position 133,225,227 to 134,111,432. The duplicated region included only a part of PCDH10. This CNV was derived from his healthy mother.
DISCUSSION
De novo KIAA2022 nonsense mutations were identified in two unrelated patients with severe ID. This report confirms that KIAA2022 mutations represent a clinically recognizable condition characterized by ID and distinctive dysmorphic features. KIAA2022 mutations and alterations have been reported in only four families with nonsyndromic ID and epilepsy [Cantagrel et al., 2004; Van Maldergem et al., 2013] (Table I). Moderate to severe ID was described in two families with frameshift mutations, one family with intron 1 disruption of KIAA2022, and the present patients with nonsense mutations. Mild ID was reported in one family with intragenic microduplication of KIAA2022, which is considered a hypomorphic phenotype. There is a correlation between the genotype and severity of ID. The consistent features among these patients were hypotonic face, narrow forehead, upturned nostrils, open mouth, and large ears. The patient with microduplication showed mild features. In this study, two of thirteen patients had KIAA2022 mutations. Significant high prevalence with these mutations is associated with selection of patients according to phenotypes and common features with KIAA2022 mutation or a chance occurrence. It is difficult to estimate the prevalence of KIAA2022 related ID.
| Van Maldergem et al., 2013 Cantagrel et al., 2004 | Van Maldergem et al., 2013 | Present patients | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Family 1 | Family 2 | Family 3 | Family 4 | |||||||
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | Patient 7 | Patient 8 | Patient 1 | Patient 2 | |
| KIAA2022 | intron 1 disruption | c.3597insA p.Ser1200Tyr*fsX5 | exon 1 70-kb duplication | c.183delC p.Arg62Glu*fsX22 | c.2113C>T p.Q705X | c.964C>T p.R322X | ||||
| Age | 13 years | 20 years | 6 years | 4 years | 8 years | 14 years | 10 years | 40 years | 3 years | 5 years |
| Walking age | 3 years | 3 years | 34 months (transient) | 4 years | 17 months | 18 months | 18 months | 14 months | Absent | 4 years |
| Language | Absent | Absent | Rudimentary | Absent | Rudimentary | Rudimentary | Poor | Poor | Absent | Absent |
| Intellectual disability | Severe to profound | Severe to profound | Mild | Moderate to severe | Severe | Severe | ||||
| Seizures | Absent | Generalized | Flexor spasms | Flexor spasms | Absent | Absent | Lennox- Gastaut | Generalized | Absent | Absent |
| Autistic behavior | +++ | +++ | +++ | +++ | + | Absent | + | Absent | +++ | +++ |
| Growth retardation | ||||||||||
| Prenatal | – | – | – | – | – | – | – | – | ||
| Postnatal | + | + | + | + | – | – | + | – | + | + |
| Microcephaly | + | – | + | + | – | – | + | – | – | + |
| Hypotonic face | + | + | + | + | + | + | + | + | ||
| Narrow forehead | + | + | + | – | + | + | + | + | ||
| Anteverted nostrils | + | + | + | + | + | + | + | + | ||
| Open mouth | + | + | + | + | + | + | + | + | ||
| Large ears | + | + | – | – | + | + | + | + | ||
KIAA2022 mutation may cause central hypothyroidism and nephrotic syndrome in addition to as ID. KIAA2022 is highly expressed in the fetal and adult brain and plays a role in neuronal development. KIAA2022 knockdown reduced the total neurite length in an in vitro study [Van Maldergem et al., 2013]. In the mouse brain, KIAA2022 expression occurred ubiquitously during the embryonic and perinatal stages and was maintained in the hippocampus, dentate gyrus, olfactory bulb, ventral thalamic nucleus, and ventromedial hypothalamic nucleus during the adult stage [Ishikawa et al., 2012]. KIAA2022 mutation is suspected to be associated with hypothalamic dysfunction, leading to central hypothyroidism in Patient 2. Xpn (mouse ortholog of KIAA2022) knockdown enhanced cell–cell and cell–matrix adhesion mediated by N-cadherin and beta 1 integrin [Magome et al., 2013]. The beta 1 integrin showed characteristic expression pattern during nephrogenesis and is crucial for normal development and function of the renal glomeruli [Kanasaki et al., 2008]. Overexpression of beta 1 integrin contributes to mesangial remodeling and is related to glomeluronephritis [Kuhara et al., 1997]. Alterations in beta 1 integrin expression pattern in location and intensity have been found in congenital nephrotic syndrome of the Finnish type [Ljungberg et al., 1996]. The occurrence of nephrotic syndrome in patient 2 suggested the possibility that dysfunction of cell adhesions could affect glomerular function due to KIAA2022 mutation. KIAA2022 is expressed in lower amounts in organ tissues other than brain, and its function in the kidney remains unclear. Other cases with KIAA2022 mutation revealed no disease or anomaly other than ID and epilepsy; there is a possibility of coincidental occurrence in Patient 2.
These additional patients support the evidence that KIAA2022 is a causative gene for X-linked nonsyndromic ID. The two patients showed consistent distinctive features and severe ID.
ACKNOWLEDGMENTS
This research was supported in part by a Grant-in-aid from the Ministry of Health, Labor, and Welfare, Japan, the Intramural Research Grant (24-8) for Neurological and Psychiatric Disorders of NCNP, and CREST, Japan Science and Technology Agency (KK). We thank the patients and their families for their co-operation.