Refinement of the deletion in 8q22.2–q22.3: The minimum deletion size at 8q22.3 related to intellectual disability and epilepsy
Abstract
Kuechler et al. [2011] reported five patients with interstitial deletions in 8q22.2–q22.3 who had intellectual disability, epilepsy, and dysmorphic features. We report on a new patient with the smallest overlapping de novo deletion in 8q22.3 and refined the phenotype. The proposita was an 8-year-old girl, who developed seizures at 10 months, and her epileptic seizure became severe and difficult to control with antiepileptic drugs. She also exhibited developmental delay and walked alone at 24 months. She was referred to us for evaluation for developmental delay and epilepsy at the age of 8 years. She had intellectual disability (IQ 37 at 7 years) and autistic behavior, and spoke two word sentences at 8 years. She had mild dysmorphic features, including telecanthus and thick vermilion of the lips. Array comparative genomic hybridization detected a 1.36 Mb deletion in 8q22.3 that encompassed RRM2B and NCALD, which encode the small subunit of p53-inducible ribonucleotide reductase and neurocalcin delta in the neuronal calcium sensor family of calcium-binding proteins, respectively. The minimum overlapping region between the present and previously reported patients is considered to be a critical region for the phenotype of the deletion in 8q22.3. We suggest that the deletion in 8q22.3 may represent a clinically recognizable condition, which is characterized by intellectual disability and epilepsy. © 2014 Wiley Periodicals, Inc.
INTRODUCTION
Rare copy number variants (CNVs) are important causes of various neurodevelopmental disorders, such as intellectual disability (ID), autism spectrum disorder (ASD), and epilepsy. Array comparative genomic hybridization (CGH) has facilitated the identification of small CNVs as candidates for ID, ASD, and epilepsy [Bartnik et al., 2012; Nicholl et al., 2013]. Kuechler et al. [2011] reported on five patients with overlapping interstitial deletions in 8q22.2–q22.3 who had moderate to severe ID and epilepsy [Kuechler et al., 2011]. In this study, we report an additional patient with the smallest overlapping deletion in 8q22.3 and we delineate the phenotype of these deletions.
CLINICAL REPORT
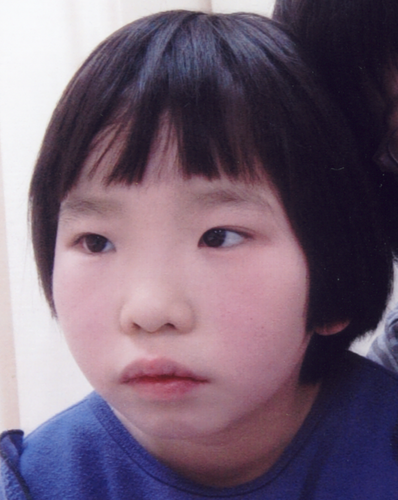
The proposita was an 8-year-old girl. She was the second child of unrelated and healthy parents. She was delivered at 39 weeks and 4 days gestational age after an uneventful pregnancy. Her birth weight was 2,702 g (−1.3 SD), length 47.5 cm (−1.2 SD), and occipital frontal circumference (OFC) 33.5 cm (0.0 SD). She started controlling her head at 5 months, sitting at 7 months, crawling at 12 months, and walked alone at 24 months. At 10 months, she developed left sided tonic clonic seizures for 30 min associated with fever. She had repeated febrile convulsive status epilepticus on four occasions and started with an angiepileptic drug at 2 years. Her epileptic seizures were severe and difficult to control with antiepileptic drugs, valproic acid, and phenobarbital. After the administration of antiepileptic drugs, she developed status epilepticus twice and had complex partial seizures with loss of consciousness, incontinence, and ocular displacement with over 10 times per day. After the administration of clobazam, she did not developed status epilepticus, but had complex partial seizures with impaired consciousness and ocular displacement with about five times per day. She was referred to us for assessment because of developmental delay and epilepsy at the age of 8 years. She had intellectual disability (IQ 37 at 7 years). She spoke first word at 3 years 10 months old and two word sentences at 8 years old. She exhibited autistic behavior and spoke stereotyped two word sentences. On physical examination at the age of 8 years, her height was 124.8 cm (−0.7 SD), weight 24.35 kg (−0.8 SD), and OFC 51.6 cm (+0.5 SD). She had mild dysmorphic features, including sparse and wide eyebrows, telecanthus, slightly flat nasal tip, and thick vermilion of the lips (Fig. 1). The electroencephalogram detected a spike and wave complex in the right frontal lobe, and generalized short burst of spike and wave. Head MRI detected sclerosis of the right hippocampus and slight atrophy of the right cerebral hemisphere.
MATERIALS AND METHODS
Written informed consent was obtained from the parents of the patient, in accordance with the Kanagawa Children's Medical Center Review Board and Ethics Committee.
Total genomic DNA was obtained from lymphocytes using standard techniques. Array CGH was performed using an Agilent SurePrint G3 Human CGH Microarray Kit 8 × 60 K (Agilent Technologies, Inc., Santa Clara, CA) covering the human genome at an overall median probe spacing of 41.5 kb, according to the manufacturer's instructions. The results were analyzed using Agilent Genomic Workbench software.
FISH analysis to determine the deletion in 8q22.3 was performed using bacterial artificial chromosome (BAC) clones, which were selected from the May 2004 (NCBI35/hg17) Assembly of the UCSC Genome Browser for Human (http://genome.ucsc.edu/). All DNA samples were labeled by nick translation, according to the manufacturer's instructions (Abbott-Vysis, Des Plaines, IL). Hybridization, post-hybridization washing, and counterstaining were performed according to standard procedures. Slides were analyzed using a completely motorized epifluorescence microscope (Leica DMRXA2), which was equipped with a CCD camera. The camera and microscope were controlled using Leica CW4000 M-FISH software (Leica Microsystems Imaging Solutions, Cambridge, UK) [Kurosawa et al., 2012].
RESULTS
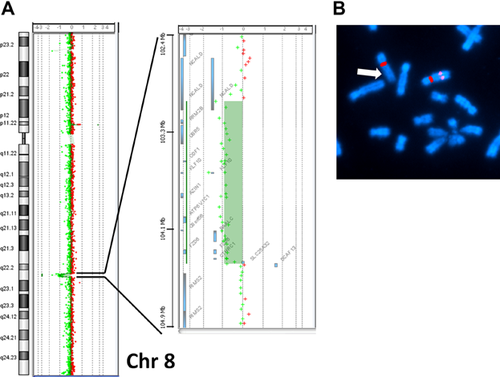
The array CGH detected an approximately 1.36 Mb deletion in 8q22.3 from position 103,066,066 to 104,430,435 bp (GRCh37/hg19 assembly). The maximal size of deleted region was from position 103,032,497 to 104,439,572 bp, estimated from the locus proximal to the deletion. The deleted region included 14 RefSeq genes, NCALD (OMIM 606722), RRM2B (OMIM 604712), UBR5 (OMIM 608413), ODF1 (OMIM 182878), KLF10 (OMIM 601878), AZIN1 (OMIM 607909), ATP6V1C1 (OMIM 603097), C8orf56, BAALC (OMIM 606602), FZD6 (OMIM 603409), CTHRC1 (OMIM 610635), SLC25A32 (OMIM 610815), DCAF13, and SLC25A32 (OMIM 610815). The deletion was confirmed by FISH analysis using the RP11–106E2 (8q22.3 chr8: 103,123,518–103,290,569) probe (Fig. 2). Neither parent had the same deletion.
DISCUSSION
We report on a new patient with the smallest deletion in 8q22.3, who had ID and epilepsy. Kuechler et al. [2011] reported on five patients with overlapping interstitial deletions in 8q22.2–q22.3 [Kuechler et al., 2011] (Fig. 3). Three patients had overlapping deletions in the ISCA database (nssv 576181, 582174, and 1494917). Two of the three patients had ID, whereas the other had ASD. They had common overlapping deletion regions in 8q22.3.

The patient refined the critical region of the deletion in 8q22.2–q22.3 with a recognizable syndrome. According to this and previous reports, patients with deletions in 8q22.2–q22.3 had consistent features, including ID, epilepsy, and a distinctive phenotype (Table I) [Kuechler et al., 2011]. The previously reported patients had moderate to severe developmental delay with autistic behavior and four of five patients had seizures. Four of the five patients (Patients 1–4) had 5–6 Mb deletions in 8q22.2–q22.3 and distinctive phenotypes, including blepharophimosis, telecanthus, epicanthus, sparse eyebrows, flat nasal tip, downturned corners of the mouth, and malformed ears. Patient 5 had a smaller overlapping deletion of 1.92 Mb in 8q22.2, with moderate developmental delay and epilepsy but no dysmorphic features, except downturned corners of the mouth. The patient also had mild dysmorphic features that differed from those of Patients 1–4. Patient 5 and the present patient revealed no recognizable phenotypes of 8q22.2–q22.3 deletion. The common deleted region (red line) is considered to be a critical region for developmental delay and epilepsy. The distal 8q22.2 or proximal 8q22.3 (green line) regions may be associated with distinctive facial phenotypes.
| Kuechler et al. [2011] | Present case | |||||
|---|---|---|---|---|---|---|
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | ||
| Deletion size | 5.26 Mb | 6.10 Mb | 5.26 Mb | 6.44 Mb | 1.92 Mb | 1.36 Mb |
| Growth retardation | ||||||
| Prenatal | – | – | – | + | – | – |
| Post-natal | – | – | + | + | – | – |
| Microcephaly | – | – | + | + | – | – |
| Intellectual disability | Severe | Moderate | Severe | Severe | Moderate | Moderate/severe |
| Sitting (months) | 12 | 24–36 | 24 | 9 | 7 | |
| Walking (months) | 27 | 17 | 60 | 90 | 24 | 24 |
| Speaking | No words | Many single words | No words | No words | 18–20 months | 3 years 10 months |
| Epilepsy | + | – | + | + | + | + |
| Autistic behavior | + | + | – | + | + | + |
| Others | – | Large hiatal hernia, pyloric stenosis, hypospadias | – | Diaphragmatic hernia | – | – |
| Blepharophimosis | + | + | + | + | – | – |
| Telecanthus | + | + | – | + | – | + |
| Epicanthus | + | + | + | + | – | – |
| Eyebrows | Sparse broad | Rather sparse | Rather sparse | Bushy | Mild | Sparse wide |
| Flat nasal tip | + | + | + | + | – | Slightly |
| Downturned corners of the mouth | + | + | + | + | + | – |
| Ear anomaly | + | + | + | + | – | – |
The minimum overlapping region in this and previously reported patients included six RefSeq genes: NCALD (OMIM 606722), RRM2B (OMIM 604712), UBR5 (OMIM 608413), ODF1 (OMIM 182878), KLF10 (OMIM 601878), and AZIN1 (OMIM 607909) (Fig. 3). Two of these genes are associated with central nervous system functions: NCALD and RRM2B. NCALD encodes neurocalcin delta in the neuronal calcium sensor family of calcium-binding proteins, which is highly expressed in brain. Wang et al. [2001] suggested that NCALD plays an important role in the regulation of the neuronal signal transduction process [Wang et al., 2001]. A member of the calcium sensor protein family, VSNL1 (MIM 600817) encoding VILIP-1 (visinin-like protein-1), is associated with Alzheimer disease and schizophrenia [Braunewell et al., 2011]. RRM2B encodes the small subunit of p53-inducible ribonucleotide reductase, which is expressed ubiquitously and plays a role in cell survival by repairing damaged DNA. Mutations in the RRM2B gene cause autosomal recessive mitochondrial DNA depletion syndrome and autosomal dominant progressive external ophthalmoplegia [Bourdon et al., 2007; Kuechler et al., 2011]. We suggest that this region encompasses candidate genes for ID and epilepsy.
In conclusion, we describe a girl with the minimum deletion size in 8q22.3 with ID and epilepsy. The findings for the patient delineated the critical regions for ID and epilepsy with a deletion in 8q22.2–22.3. The candidate gene for ID and epilepsy may be involved with the minimum overlapping region.
ACKNOWLEDGMENTS
This research was supported in part by a Grant-in-aid from the Ministry of Health, Labor, and Welfare, Japan and an Intramural Research Grant (24–8) for Neurological and Psychiatric Disorders from NCNP. We thank the patient and the family for their cooperation.