Characterization of a 520 kb deletion on chromosome 15q26.1 including ST8SIA2 in a patient with behavioral disturbance, autism spectrum disorder, and epilepsy
Abstract
We present a patient with a behavioral disorder, epilepsy, and autism spectrum disorder who has a 520 kb chromosomal deletion at 15q26.1 encompassing three genes: ST8SIA2, C15orf32, and FAM174B. Alpha-2,8-Sialyltransferase 2 (ST8SIA2) is expressed in the developing brain and appears to play an important role in neuronal migration, axon guidance and synaptic plasticity. It has recently been implicated in a genome wide association study as a potential factor underlying autism, and has also been implicated in the pathogenesis of bipolar disorder and schizophrenia. This case provides supportive evidence that ST8SIA2 haploinsufficiency may play a role in neurobehavioral phenotypes. © 2013 Wiley Periodicals, Inc.
INTRODUCTION
Alpha-N-Acetyl-neuraminide alpha-2,8-sialyltransferase 2 (ST8SIA2, SIAT8B, or STX) encodes a golgi-resident enzyme which catalyzes the post-translational transfer of sialic acid onto substrate proteins, and is principally responsible for glycosylation of the neural cell adhesion molecule 1 (NCAM1) [Finne, 1982; Eckhardt et al., 1995]. In its polysialylated form (PSA-NCAM), it plays an important role in processes such as neuronal migration and axon pathfinding, synaptogenesis and plasticity [Finne, 1982; Eckhardt et al., 1995; Nakayama et al., 1995; reviewed in Rutishauser, 2008; Muhlenhoff et al., 2013]. It is highly expressed during early brain development and expression decreases with age, and becomes restricted to areas of neurogenesis and neuroplasticity in adults [Hildebrandt et al., 2009; McAuley et al., 2009].
Homozygous St8sia2-deficient mice had unremarkable physical and metabolic assessments and were not impaired in spatial learning using the water maze test. However, St8sia2-deficient mice exhibited higher exploratory drive and reduced behavioral responses to Pavlovian fear conditioning. These mice exhibited misguidance of infrapyramidal mossy fibers and formation of ectopic synapses in the hippocampus. Data was not presented for heterozygous mice [Angata et al., 2004].
Genome wide association studies have provided evidence that variants in or near ST8SIA2 are associated with autism and bipolar disorder [Anney et al., 2010; Lee et al., 2011]. Earlier genome wide linkage studies implicated the 15q26 region in schizophrenia and bipolar disorder and a recent fine mapping association study has implicated a specific risk haplotype in ST8SIA2 with increasing genetic risk in both schizophrenic and bipolar patients [Maziade et al., 2005; Arai et al., 2006; Tao et al., 2007; Vazza et al., 2007; McAuley et al., 2009; McAuley et al., 2012].
Here we present a child with epilepsy and autism spectrum disorder with a severe behavioral profile with a 520 kb deletion of three genes on 15q26.1, including ST8SIA2.
CLINICAL REPORT
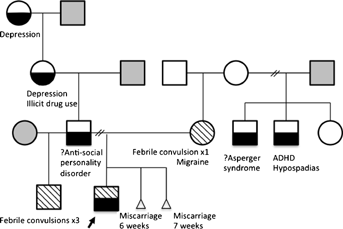
The patient was seen at age 5 years, 9 months. He is the first child to nonconsanguineous parents. We did not have the opportunity to meet his father who has a diagnosis of antisocial personality disorder (we could not access his medical records to confirm this). There is a strong family history of depression on the father's side of the family. The proband has a paternal half-brother who had three febrile convulsions. Information regarding the half-brother's intellectual functioning and behavior is not available. The patient's mother suffers migraines and is intellectually normal. She has a half-brother with attention deficit hyperactivity disorder (ADHD) and hypospadias, and another half-brother with Asperger like traits, although no formal diagnosis has been made. The pedigree is shown in Figure 1. The pregnancy history was unremarkable. Our patient was born at 38 weeks gestation via cesarean for cephalopelvic disproportion. There were no neonatal complications.
The patient has epilepsy that started at age 6 months. He has had presumed absence seizures, drop seizures, and generalized tonic clonic seizures. These occur every few days to every few months. He had three febrile convulsions between 16 and 24 months.
Behavioral abnormalities in the proband include hyperactivity, impulsivity, poor attention, frequent tantrums, occasional aggression, and a fixation with sharp objects. He occasionally threatens self-harm or violence to others. He is over-familiar with strangers and can exhibit inappropriate hugging or kissing. Social reciprocity is poor. For example, he will frequently go very close to other children's faces and “roar,” or push them over. He also exhibits repetitive play and repetitive speech and dislikes changes to his routine. From birth, he exhibited constant crying and poor sleep. His poor sleep continues requiring melatonin treatment. He also has a high pain threshold. Developmental milestones such as walking and speech were attained at appropriate times, but his mother reports that he was never interested in people. She describes his behavior as unusual from a young age, including pica.
At age 6 years, 9 months, the Autism Diagnostic Observational Schedule (ADOS 2) and the Autism Diagnostic Interview (ADI-R) were administered. The patient met criteria for autism spectrum disorder based on ADOS 2, ADI-R, and DSM-V criteria, respectively. The results of the ADOS 2 and ADI-R are listed in Table I.
| Scores | |
|---|---|
| ADOS 2 | |
| Social affect total | 20 |
| Restricted and repetitive behaviors total | 5 |
| Social affect and restricted and repetitive behaviors overall total | 25 (cut off score = 9) |
| ADI-R | |
| Qualitative abnormalities in reciprocal social interaction | 23 (cut off score = 10) |
| Qualitative abnormalities in communication | 18 (cut off score = 8) |
| Restricted, repetitive and stereotyped patterns of behavior | 7 (cut off score = 3) |
| Abnormality of development evident at or before 36 months | 3 (cut off score = 1) |
- The numbers in brackets indicate cut off scores. A score above the cut off suggests a diagnosis of autism.
Formal developmental assessment was performed at age 5 using the Wechsler Preschool and Primary Scale of Intelligence (3rd Edition; Australian Standardized Edition). Due to frustration and aggression, a complete assessment was not possible although our patient was estimated to have a verbal IQ in the average range (27th centile) and a performance IQ in the low average range of abilities (14th centile).
The patient has been treated pharmacologically with topiramate, carbemazepine, and melatonin. A previous trial of sodium valproate was associated with a significant decline in behavior, by parental report. Changing to topiramate, his mother noticed an almost immediate improvement in behavior and de-escalation of aggression and violence.
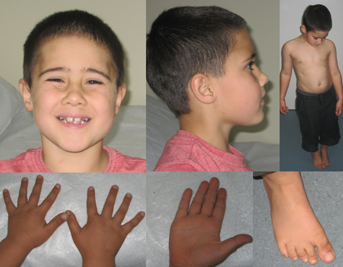
Physical examination revealed a boy with a head circumference of 51.2 cm (50th centile). His height was on the 75th centile and weight on the 90th centile. He had mild eczema. Photographs are shown in Figure 2.
Brain imaging investigations have included a cerebral MRI, which was within normal limits at age 5. Electroencephalograms have been normal or showed isolated theta waves in the right frontal region. Fragile X testing, urine metabolic screening, plasma amino acids, ammonia, electrolyte analysis, lead levels, creatine kinase level, and full blood count analysis were unremarkable.
MATERIALS AND METHODS
Molecular analysis used an ISCA 60k genome-wide comparative genomic hybridization (CGH) array version 2 analyzed with BlueFuse-Multi software (BlueGnome, Fulbourn, Cambridge, UK).
The DECIPHER database was used to view and interpret the chromosomal microdeletion. Haploinsufficiency indexes taken from the DECIPHER datatabase were used. Low scores indicate a high predicted probability that a gene is haploinsufficient, that is, that deletion of one allele may have a pathogenic effect [Firth et al., 2009; Huang et al., 2010].
Sequencing of the transcribed and regulatory regions of ST8SIA2 was conducted following standard v3.1 Big-Dye terminator sequencing methods, using the ABI377 automated sequencer (Life Technologies, Mulgrave, Victoria, Australia). Traces were base-called and visualized using phred, phrap, and consed software [Ewing et al., 1998; Gordon et al., 1998].
Haplotypes were phased using PLINK software [Purcell et al., 2007], augmented with genotype data from 176 Caucasian European Individuals downloaded from 1000 Genomes Data Browser [Abecasis et al., 2012]. Alleles at each SNP were defined as reference or alternative, according to those annotated in UCSC [Kent et al., 2002; Meyer et al., 2013]. Base positions are all stated relative to the NCBI build 36/hg18 (March 2006 release) genome assembly.
RESULTS
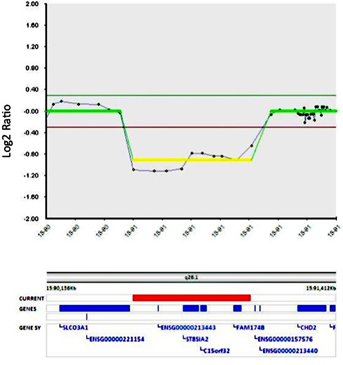
We detected a heterozygous deletion at chromosome 15q26.1 via array CGH, spanning 520 kb (chr15:90,517,962–91,039,825; NCBI36/hg18). The three genes in this region include ST8SIA2, C15orf32, and FAM174B (Fig. 3). The deletion was confirmed through FISH analysis with the probe RP11-304N14 (chr15: 90,821,315–90,929,683). We found no evidence of this specific deletion from previous reports within the DECIPHER database (see the Discussion Section for more information regarding overlapping CNVs in DECIPHER and other databases) [Firth et al., 2009]. The patient's mother does not have this deletion, as tested by FISH analysis. Unfortunately, we have not been able to arrange testing of the patient's father.
Direct sequencing of the transcribed and 5′ regulatory region of ST8SIA2 was conducted in the DNA from the patient and his mother. This comprised a total of 12.7 kb, including around 450 bp surrounding exons 2–5, plus 5.2 kb of promoter 5′UTR and exon 1 sequence (chr15:92,933,453–92,938,739), and 5 kb of exon 6 comprising both coding and 3′UTR (chr15:93,007,253–93,012,218). A total of 218 known SNPs (as annotated in dbSNP137) were genotyped: the mother was homozygous for 207 SNPs (with UCSC reference allele in 195 SNPs, alternative allele in 12 SNPs) and heterozygous for 11 SNPs which were distributed throughout the gene. The patient was hemizygous at all SNPs, consistent with the CGH array results of a deletion. He had identical genotypes to his mother, with the exception of the 11 maternal heterozygous SNPs, where he had inherited the reference allele (as defined by UCSC) at 6 SNPs and the alternative allele at 5 SNPs. In total, he had 17 SNPs with non-reference alleles. The non-deleted copy in the patient contains no variation in the coding or regulatory sequence which has not been described previously (dbSNP), and all variations within the protein coding sequence are UCSC reference alleles.
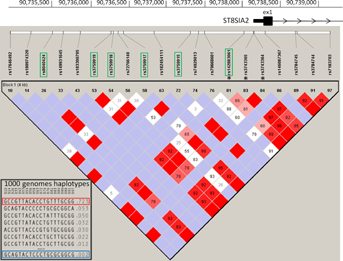
Haplotype analysis of SNPs in the promoter/exon1 region (chr15:90,734,862–90,739,345), which included three SNPs which were heterozygous in the mother and 13 out of the total 17 non-reference alleles identified in the son, shows that the single haplotype present in the patient is rare in the general population (freq = 0.0024; Fig. 4). This rare haplotype is present in the patient's mother, along with the most common haplotype (freq = 0.723). Neither of these maternal haplotypes are represented by the risk or protective haplotypes previously reported to be associated with bipolar disorder and schizophrenia [McAuley et al., 2012]. The rarity of the remaining haplotype in the patient suggests the deletion event may have arisen on the paternal chromosome, either through direct inheritance or a de novo mutational event, although DNA from the father would be required to confirm this.
DISCUSSION
We have studied a patient with a heterozygous 520 kb deletion at 15q26.1, spanning three genes: ST8SIA2, C15orf32 and FAM174B. No similar deletions have been detected in databases of normal individuals, represented in the Database of Genomic Variants (DGV) and the Children's Hospital of Philadelphia (CHOP) database. There are 14 reports of individuals with larger CGH abnormalities of the 15q26 region in the DECIPHER database although only five losses (size range 2.29–14.9 Mb) [Firth et al., 2009]. There are also three reports of patients with small gains of the equivalent region including ST8SIA2, C15orf32, and FAM174B (<500 kb), one had aggressive behavior, brachycephaly, clinodactyly, coarse hair, intellectual disability/developmental delay, and microcephaly. Phenotypic information for the other patients was not supplied to the DECIPHER database. According to the DECIPHER database, our patient has the smallest reported microdeletion involving ST8SIA2.
Larger deletions of the 15q26 region have been associated with congenital diaphragmatic hernia (CDH) and a critical region spanning ∼5 Mb on 15q26.1–26.2 (90.635–96.342 Mb), including ST8SIA2 and six distal (telomeric) genes has been identified [Biggio et al., 2004; Klaassens et al., 2005]. While our patient has a deletion which lies within the CDH critical region, our patient has no clinical similarities to those typically shared by CDH patients. There is one reported patient with a larger 3.3 Mb deletion including ST8SIA2 and 17 other mostly proximal genes (89.185–92.511 Mb) chr15:90,734,862–90,739,345 [Li et al., 2008]. At age 5 he had developmental delay, mild dysmorphic features and a history of febrile convulsions. Further, a patient with a 4.9 Mb deletion including ST8SIA2 and ∼40 other mostly proximal genes (87.796–92.700 Mb) [Veredice et al., 2009] suffered early onset refractory myoclonic epilepsy, intellectual disability and growth delay.
Although no CNV variations were identified in phase I of the Autism Genome Project affecting ST8SIA2, C15orf32, or FAM174B, common single nucleotide polymorphisms in ST8SIA2 have previously shown genome-wide significant association with autism spectrum disorder (verbal subtype) [Anney et al., 2010; Pinto et al., 2010]. This indicates that while ST8SIA2 may contain common variation that increases risk for autistic traits, heterozygous deletion of this gene appears to be rare in patients with autism. However, given the previous association of ST8SIA2 with autism spectrum disorder, its involvement in the orchestration of early brain neurodevelopment and plasticity, and the very early onset nature of some behaviors in our patient, we hypothesize that haploinsufficiency of ST8SIA2 contributes to the phenotype of our patient [Rutishauser, 2008; Anney et al., 2010].
Neither C15orf32 nor FAM174B are OMIM-listed, and there is little published about their function. According to the Unigene and CGAP datasets (accessed via Genecards), C15orf32 does not appear to be expressed in the brain. According to DECIPHER, its haploinsufficiency index is 90.4% making this gene an unlikely candidate for behavioral abnormalities when heterozygously deleted [Firth et al., 2009; Huang et al., 2010]. FAM174B is a membrane protein, which is expressed in the brain according to Unigene, CGAP, and BioGPS datasets. A haploinsufficiency index has not been formulated. This deleted gene could contribute to the proband's phenotype.
The haploinsufficiency index for ST8SIA2 is 26.6%, indicating that ST8SIA2 loss-of-function could lead to deleterious effects [Huang et al., 2010]. Transgenic studies in mice have shown that presence of a single copy of st8sia2 is capable of polysialylating ∼90% of available NCAM, and is sufficient to prevent the severe brain morphological defects seen in mutant mice who were polySia-negative and NCAM positive [Galuska et al., 2006; Hildebrandt et al., 2009]. We were able to confirm that no additional mutations affecting the coding region of ST8SIA2 are present, which would reduce the functionality of the remaining allele or completely knockout ST8SIA2 in our patient. Previous studies in mice have shown severe phenotypes in homozygous mutant mice, and in mice with compound mutations [Angata et al., 2004; Hildebrandt et al., 2009]. Consistent with the studies in heterozygous transgenic mice, no gross morphological changes were observed in the brain MRI of our patient.
Traditionally, behavioral disorders are considered to be multifactorial and polygenic [Dick, 2011; Schaaf et al., 2011; Leblond et al., 2012; Viding et al., 2013]. While no other large duplications or deletions were identified in our patient, it is likely that variants in other genes contribute to the observed phenotype. There may also be other (perhaps as yet unknown) regulatory or modifying loci related to ST8SIA2 that contain contributory variants.
Based on the haplotype of the remaining ST8SIA2 allele in the patient, it appears that the deletion itself was not inherited from the mother. However, a major limitation of this report is that we were unable to determine whether the microdeletion occurred de novo, or was inherited from a father who may also have a behavioral phenotype. In any case, it is likely there is genetic contribution from both sides of the family as a maternal half uncle has attention deficit hyperactivity disorder and another maternal half uncle has Asperger-like traits.
Behavioral issues are common and often related to difficult life circumstances and poor attachment. Our patient was in a chaotic home environment for the first 18 months of life, but has had no further contact with his father and a more stable home life since. It is possible that his behavior is purely a circumstance of his early environment. Epilepsy (particularly with frontal lobe involvement) can also impact significantly on behavior, and this is another potential contributing factor underlying his behavioral phenotype.
Despite the other potential genetic and environmental causes or contributions, this report provides evidence that ST8SIA2 haploinsufficiency may cause or create a susceptibility to various conditions such as behavior disorder, autism spectrum disorder and epilepsy. Of course, caution is needed when drawing conclusions from a single patient and we await further reports of other similarly affected patients.
WEB RESOURCES
- DECIPHER database, http://decipher.sanger.ac.uk/
- Genecards, http://www.genecards.org/
- 1000 Genomes Data Browser, http://browser.1000genomes.org/index.html/
- UCSC, http://genome.ucsc.edu/
- Database of Genomic Variants, http://projects.tcag.ca/variation/
- Children's Hospital of Philadelphia database, http://cnv.chop.edu/
ACKNOWLEDGMENTS
We would like to thank Monica Wordsworth (Clinical Psychologist) for the formal developmental assessment of the patient, and Dr Anand Naidoo (General Pediatrician) for referring the patient. We would also like to thank our patient and his mother who kindly allowed participation and information for this paper. The molecular work was supported by National Health and Medical Research Council grants 630574 and 1037196.




