Severe intellectual disability, West syndrome, Dandy–Walker malformation, and syndactyly in a patient with partial tetrasomy 17q25.3
Abstract
We report on a de novo 0.5 Mb triplication (partial tetrasomy) of chromosome 17q25.3 in a 10-year-old girl with severe intellectual disability, infantile seizures (West syndrome), moderate hearing loss, Dandy–Walker malformation, microcephaly, craniofacial dysmorphism, striking cutaneous syndactyly (hands 3–4, feet 2–3), joint laxity, and short stature. The triplication resulted from the unusual combination of a terminal duplication at 17qter and a cryptic translocation of an extra copy of the same segment onto chromosome 10qter. The breakpoint at 17q25.3 was located within the FOXK2 gene. SNP chip analysis suggested that the rearrangement occurred during paternal meiosis involving both paternal chromosomes 17. © 2013 Wiley Periodicals, Inc.
INTRODUCTION
Partial trisomy 17q is rare. About 30 reports in live born infants have been published so far [reviewed by Sarri et al., 1997; Schinzel, 2001; Kelly et al., 2002; Lukusa and Fryns, 2010]. The majority of duplications involve the distal third of 17q (17q23-qter) with variable breakpoints from 17q21 to 17q25.3 [Lukusa and Fryns, 2010]. Despite different extensions of the trisomic segment, the phenotype of a “17q duplication syndrome” has been defined [Naccache et al., 1984; Schinzel, 2001]. Most observations of this phenotype have been made in patients with inherited or de novo translocations involving partial trisomy 17q together with an imbalance for another chromosomal segment (i.e., double segment imbalances) [reviewed by Sarri et al., 1997; Lukusa and Fryns, 2010], and only a few patients have been recorded with a pure duplication [Orye and Van Bever, 1985; Serotkin et al., 1988; Shimizu et al., 1988; Lukusa and Fryns, 2010].
To our knowledge, tetrasomy of 17qter has not been reported so far. We describe a 10-year-old girl with a de novo partial tetrasomy 17q25.3 and compare the clinical findings with those that have been observed in patients with partial trisomy of the distal portion of 17q.
CLINICAL REPORT
The proposita was the only child of healthy, nonconsanguineous, German parents aged 38 years (mother) and 46 years (father). During the last trimester of pregnancy, gestational diabetes was treated with insulin. The mother also reported that the pregnancy was complicated by oligohydramnios and hydrops. Reports on prenatal ultrasound were not available.
The child was born at term by vaginal delivery. Birth measurements were within the upper normal range (length 53 cm, weight 3,970 g). Generalized muscular hypotonia and infantile spasms were noted during the first months of life, indicating a severe neurological disorder. Electroencephalogram (EEG) showed hypsarrhythmia which lead to the diagnosis of West syndrome. Seizures were refractory to antiepileptic drugs and pyridoxine. During a therapeutic trial with adrenocorticotropic hormone (ACTH) the frequency of spasms diminished but complete remission was not achieved.
Cranial magnetic resonance imaging (MRI) in early childhood showed a Dandy–Walker malformation and a general delay in white matter myelinization. Latency of visual evoked cortical potentials (VECP) was initially found to be increased, but was normal at the age of 3 years. Brainstem audiometry showed moderate hearing loss (below 40 dB). Due to recurrent urinary infections cystomanometry was performed at the age of 2 years revealing detrusor overactivity and detrusor/sphincter dyssynergy which was treated by an anticholinergic drug (propiverine) and intermittent catheterization. Gastro-esophageal reflux was present from early on but improved during childhood. Oropharyngeal and genital inspection, ultrasound of internal organs, and spinal MRI showed normal results.
At the age of 10 years the girl had severe global developmental delay. Unsupported sitting was not possible and she was unable to stand or walk. Her communicative skills were very limited. She had poor eye contact and severe speech impairment with no use of words. She did not use gestures. The parents were able to recognize different vocalizations or behaviors that signaled different needs. The parents reported auto-aggressive tendencies such as self-biting.
At the age of 10 years the patient still had generalized muscular hypotonia, athetoid movements (no spasticity) and pronounced laxity of the large joints. She was microcephalic (OFC = 49 cm; 1.2 cm below 3rd centile) and her stature was short (length = 110 cm; 16 cm below 3rd centile). She had alternating strabismus. Dysmorphic signs included bitemporal narrowing, deep set eyes, broad nose, anteverted nares, smooth philtrum, prominent maxilla, broad mouth with downturned corners, small mandible, and malformed low-set but normal-sized ears (25th centile), unusual palmar flexion creases, broad and clubbed nails (more pronounced on fingers), and striking cutaneous syndactyly of fingers 3–4 and toes 2–3 (for details, see Fig. 1).
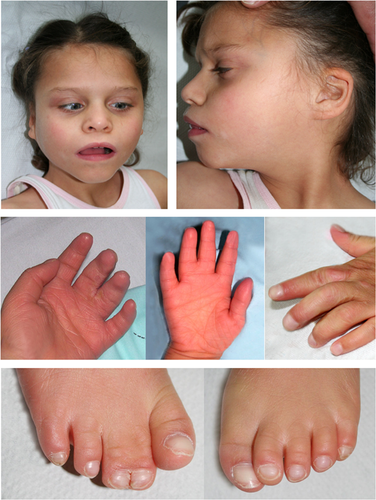
Laboratory analyses including organic acids, amino acids, orotate, sterols, sulfite test, thyroid hormones, and genetic tests for Rett- and Angelman-syndrome (sequencing and MLPA (Multiplex Ligation-dependent Probe Amplification) of MECP2, and methylation-specific PCR of SNRPN) showed normal results. Conventional karyotyping of lymphocytes (30 metaphases, ISCN band level 400–550) and of cultured skin fibroblasts (30 metaphases, ISCN band level 400) showed a normal female karyotype 46,XX. There was no hint of a diploid/triploid mosaicism by fluorescence in situ hybridization (FISH) on 50 metaphases from skin fibroblasts (PrenatScreen kit, KREATECH).
METHODS
Array CGH (comparative genomic hybridization) analysis was performed using the standard Human Genome CGH Microarray Kit 244A (Agilent, Santa Clara, CA) according to the manufacturer's protocols. After hybridization arrays were scanned with the Agilent microarray scanner, raw data were processed with the Feature Extraction 9.5.3.1 software (Agilent) and analyzed with the DNA Analytics 4.0.76 program (Agilent). For validation and fine mapping of breakpoints a customized array CGH Chip (OGT, Oxford, UK) was used. Generation of lymphoblastoid cell lines, cell culture, and fluorescence in situ hybridization (FISH) experiments were performed according to standard protocols. BACs covering regions on 17qter (RP11-388C12), and 10qter (RP11-297K12, RP11-165M8) were from Source Bioscience (Cambridge, UK). According to the UCSC map, BAC RP11-388C12 is the most telomeric clone on 17q25.3 and BAC RP11-297K12 is the most telomeric clone on 10q26.3. Clone RP11-165M8 which maps to 10q23 (PTEN) was used as hybridization control.
MLPA was performed according to the manufacturer's description, using the SALSA MLPA kit P320-A1 human telomere-13 (MRC Holland, Amsterdam, Netherlands) and a control DNA pooled from eight individuals. MLPA testing was performed twice on two different DNA samples of the patient.
SNP array analysis with the HumanCytoSNP-12v2.1 Bead Chip (Illumina, San Diego, CA) was performed according to the manufacturer's instructions. This array contains 299.140 single nucleotide polymorphism (SNP) markers with a mean intermarker distance of about 10 kb. Raw SNP calling data were processed using the Genotyping Analysis Module of GenomeStudio 1.6.3 (Illumina). Parent of origin was determined by comparison of proband's and parental genotypes and B-allele frequencies for SNPs in the triplicated region on chromosome 17.
The DNA copy number gain reported here has been entered into the DECIPHER database (#263035).
Genomic Rearrangement
Oligonucleotide array CGH analysis detected a 0.5 Mb DNA copy number gain 17q25.3–17qter (Fig. 2). A log2(ratio) of about 1 indicated that four copies of this region were present (i.e., with two reference alleles, log2(4/2) = 1). The triplicated region contained seven MIM-recorded genes (ZNF750, TBCD, FN3KRP, FN3K, RAB40B, WDR45L, and FOXK2). The proximal breakpoint of the segment was assigned to position 80.542–80.548 Mb (hg19, Fig. 2), disrupting the FOXK2 gene. In order to determine the structural basis of the triplication BAC RP11-388C12 mapping to 17qter was used for FISH on metaphase spreads and interphase nuclei. Analysis of 100 metaphase spreads from a lymphoblastoid cell line showed an enhanced signal on one chromosome 17, a signal of normal intensity on the other homologue, and an additional signal in the q-terminal region of one chromosome 10 (Fig. 3A). Interphase FISH of 100 lymphocyte as well as 100 fibroblast nuclei revealed at least three (one of them being enhanced; Fig. 3C) or, in some cases, four signals (Fig. 3D) in each nucleus.

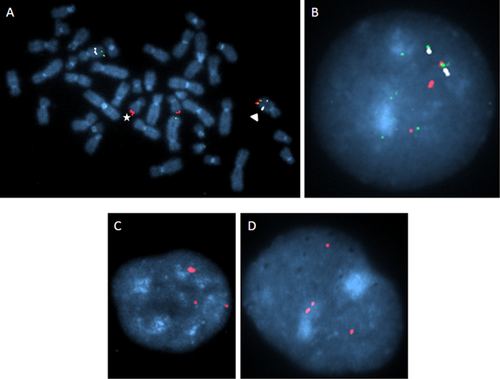
Simultaneous hybridization of BAC RP11-297K12 (chromosome band 10q26.3) and BAC RP11-388C12 (chromosome band 17q25.3) on metaphase spreads showed that the translocated segment of chromosome 17 was attached to the terminal band of 10q (Fig. 3A,B) without a copy number loss on 10q as indicated by array CGH.
Customized array CGH and FISH analyses of the parents showed normal results (data not shown). Standard array CGH indicated an additional small (0.06 Mb) heterozygous deletion at 16q23 which was inherited from the mother. The deletion itself did not seem to be pathogenic because the mother did not show any of the pathological features of her daughter.
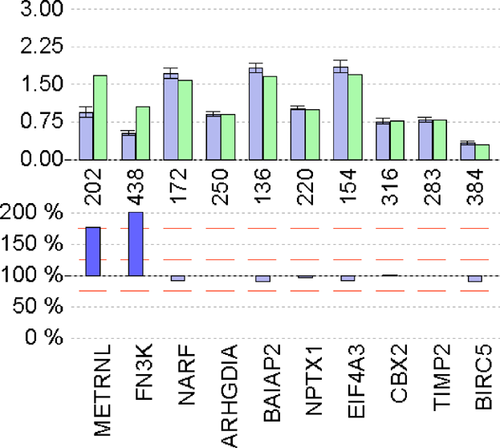
MLPA confirmed the array CGH finding of partial tetrasomy 17q. For the two distal genes METRNL and FN3K which map to 17q25.3, the relative peak area (RPA) ratios were 176.3 ± 4.4 (mean ± SD in %) and 191.9 ± 9.6, respectively, indicating a fourfold copy number. The RPA ratio for the proximally located NARF gene was normal (93.1 ± 2.1) indicating that the breakpoint was located between NARF at 80.42 Mb and FN3K at 80.69 Mb which was in keeping with the array CGH findings (Fig. 4).
In order to obtain information on the parental origin of the tetrasomy we performed SNP array analysis of the patient's and the parents' DNA. Pattern of B-allele frequency was in line with tetrasomy 17q25.3. Comparison of the patient's with the parental genotypes for 12 informative SNPs (Table I) indicated that the rearrangement most probably arose in the father during meiosis involving both paternal alleles. Alternative explanations (e.g., origin in maternal meiosis or a postzygotic origin) were not consistent with the SNP data. However, due to the inherent uncertainty of genotype calls, a postzygotic origin cannot completely be excluded.
| SNP | Mother | Father | Child |
|---|---|---|---|
| rs3794725 | BB | AB | ABBB |
| rs2279395 | AB | BB | ABBB |
| rs2250754 | AA | AB | AABB |
| rs4986122 | AB | BB | ABBB |
| rs1078334 | AA | AB | AAAB |
| rs3785520 | AB | BB | ABBB |
| rs9303016 | BB | AB | AABB |
| rs731447 | AA | AB | AABB |
| rs6502033 | AA | AB | AABB |
| rs7406132 | AA | BB | ABBB |
| rs9908845 | AB | AB | AAAB |
| rs6502042 | AA | AB | AABB |
DISCUSSION
Here, we present a patient with a pure 0.5 Mb triplication of 17q25.3-qter which is to the best of our knowledge the first report on partial 17q tetrasomy in the literature.
Partial trisomy of the long arm of chromosome 17 is regarded as a defined clinical entity [Naccache et al., 1984; Sarri et al., 1997; Schinzel, 2001]. Common findings include developmental delay and intellectual disability of varying degrees, short stature, microcephaly, various dysmorphisms, skeletal abnormalities such as rhizomelia, polydactyly or syndactyly of fingers/toes, and joint laxity. Malformations of the heart, the kidneys, and the central nervous system (CNS) ranging from Dandy–Walker malformation to holoprosencephaly are also frequent. Characteristic cranio-facial features are a narrow prominent forehead, flat nasal bridge, broad nose, wide mouth with downturned corners and a thin vermilion of the lips, high or cleft palate, small mandible, low-set and dysplastic ears, and a low posterior hairline [Sarri et al., 1997; Lukusa and Fryns, 2010]. Lukusa and Fryns [2010] recently reported on a patient with a dup(17)(q25.3qter) of 0.6–2.5 Mb who presented with growth retardation, developmental delay, distal arthrogryposis, and characteristic dysmorphic features of dup(17q) syndrome. The authors stated that arthrogryposis is an unusual finding in patients with duplication 17q and discussed that it resulted from fetal akinesia of their patient. They reviewed five cases with pure duplication 17q and concluded that the critical region for the majority of the features in dup(17q) syndrome is not as large as the region 17q23-17qter which was previously suggested [Naccache et al., 1984; Sarri et al., 1997] but is confined to chromosome bands 17q25.3-17qter. This hypothesis is corroborated by the patient presented here who shows all the typical features of trisomy 17q syndrome, that is, intellectual disability, short stature, microcephaly, bitemporal narrowing, wide mouth with down-turned corners, broad nose, malformed ears, syndactyly, joint laxity, and CNS abnormalities. Incidentally, there were features (deep set eyes, strabismus, and repetitive hand movements) shared by our patient and the patient reported by Lukusa and Fryns [2010]. These features were so far not known to be typical for dup(17q) syndrome but may now be regarded as part of the clinical spectrum of this chromosome disorder.
Despite the significant clinical overlap with dup(17q) syndrome the specific effects of 17q triplication on gene expression and on the phenotype still remain to be resolved. In principle, the effect of a triplication of 17q could simply be stronger and lead to a more severe phenotype but it could also have a qualitatively different impact on the clinical outcome.
We cannot exclude that the disruption of the FOXK2 gene at the 17q breakpoint also contributes to the phenotype. Moreover, the gene regulation of the translocated 17q-segment on chromosome 10 might be influenced by the altered genomic environment. Nevertheless, the present case may help to refine the genotype–phenotype map of 17q copy number gains.
Some of our patient's features (growth delay, syndactyly, microcephaly, cerebral malformation, and intellectuall disabillity) overlap with Filippi syndrome (OMIM %272440), an autosomal-recessively inherited disorder of so far unknown origin [Battaglia et al., 2008].
The majority of partial tetrasomies occurs as a result of isochromosome or isodicentric chromosome formation. Moreover, a small number of cases with intrachromosomal triplications have been reported [reviewed in Rivera et al., 1998; Burnside et al., 2009]. To our knowledge, partial tetrasomies resulting from duplication plus translocation as observed in the present patient have not been described before.
SNP array analysis allowed us to get more insight into the mechanism of formation of this rearrangement. Most probably, the rearrangement arose during paternal meiosis involving both paternal chromosomes 17. As such, the rearrangement may have been linked to a crossing-over event.
In conclusion, our observation demonstrates that for precise analysis of a genomic multiplication event, array CGH needs to be complemented by other molecular cytogenetic examinations, and it shows that chromosome aberrations may turn out to be more complex than initially expected.




