MLL2 and KDM6A mutations in patients with Kabuki syndrome
Abstract
Kabuki syndrome is a congenital anomaly syndrome characterized by developmental delay, intellectual disability, specific facial features including long palpebral fissures and ectropion of the lateral third of the lower eyelids, prominent digit pads, and skeletal and visceral abnormalities. Mutations in MLL2 and KDM6A cause Kabuki syndrome. We screened 81 individuals with Kabuki syndrome for mutations in these genes by conventional methods (n = 58) and/or targeted resequencing (n = 45) or whole exome sequencing (n = 5). We identified a mutation in MLL2 or KDM6A in 50 (61.7%) and 5 (6.2%) cases, respectively. Thirty-five MLL2 mutations and two KDM6A mutations were novel. Non-protein truncating-type MLL2 mutations were mainly located around functional domains, while truncating-type mutations were scattered through the entire coding region. The facial features of patients in the MLL2 truncating-type mutation group were typical based on those of the 10 originally reported patients with Kabuki syndrome; those of the other groups were less typical. High arched eyebrows, short fifth finger, and hypotonia in infancy were more frequent in the MLL2 mutation group than in the KDM6A mutation group. Short stature and postnatal growth retardation were observed in all individuals with KDM6A mutations, but in only half of the group with MLL2 mutations. © 2013 Wiley Periodicals, Inc.
INTRODUCTION
Kabuki syndrome (KS; OMIM 147920) is a multiple congenital anomaly syndrome that was originally reported by Niikawa et al. [1981] and Kuroki et al. [1981] (also known as Kabuki make-up syndrome or Niikawa–Kuroki syndrome). KS is diagnosed clinically by characteristic facial features, including long palpebral fissures and ectropion of the lateral third of the lower eyelids, postnatal growth impairment (short stature), developmental delay, intellectual disability, dermatoglyphic abnormalities, visceral and skeletal abnormalities, and immunological dysfunction. The prevalence of the disorder is estimated to be 1 in 32,000 live births [Niikawa et al., 1988]. Two genes have shown to be mutated in patients with KS: MLL2 (myeloid/lymphoid or mixed-lineage leukemia 2; NM_003482.3) at 12q13.12 and KDM6A (lysine (K)-specific demethylase 6A; NM_021140.2) at Xp11.3 [Ng et al., 2010; Lederer et al., 2012; Miyake et al., 2013]. MLL2 encodes a histone H3 lysine 4 (H3K4)-specific methyl transferase and KDM6A is a specific demethylase of histone H3 lysine 27 (H3K27) [Prasad et al., 1997; Lee et al., 2007]. They are both trithorax group proteins and bind each other [Schuettengruber et al., 2007]. These proteins are important for the chromatin state and transcription activation: MLL2 methylates H3K4 and KMD6A removes the H3K27 trimethylation repressive mark [Dubuc et al., 2013]. The loss of MLL2 or KDM6A function may lead to repressed transcription [Dubuc et al., 2013].
To our knowledge, there has been no comprehensive screen for mutations in these two genes in the same patient series. In this report, we performed a mutation screen of both genes in 81 patients with KS. We then evaluated the clinical features based on the genetic information.
MATERIALS AND METHODS
Samples
Eighty-one individuals clinically suspected to have KS were incorporated in this study: 77 Japanese, two Caucasians, one Belgian, and one Thai. They were all sporadic except for KMS-79, who had an affected sibling. Peripheral blood samples or saliva samples from the patients and their parents (when available) were collected with informed consent and DNA was extracted using a QuickGene-610L (Fujifilm, Tokyo, Japan) or Oragene-DNA kit (DNA Genotek, Inc., Ottawa, Canada) according to the manufacturer's instructions. This study included four previously reported patients (KMS-50, KMS-51, KMS-61, and KMS-71) [Tekin et al., 2006; Torii et al., 2009; Ito et al., 2013]. In addition, three patients with a KDM6A mutation were previously described as Patients 1, 2, and 3 by Miyake et al. [2013], and are named KMS-31, KMS-37, and KMS-65, respectively, in this report. This study was approved by the Institutional Review Board of Yokohama City University School of Medicine.
Mutation Screening
Fifty-eight patients (KMS-01 to KMS-69) were screened for MLL2 mutations by the high-resolution melting (HRM) method using a LightCycler 480 System II (Roche Diagnostics, Indianapolis, IN) and subsequent Sanger sequencing. If an HRM curve pattern was different from those of controls, the DNA sample was Sanger sequenced on an ABI 3500xl or 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA) and the sequences were analyzed using Sequencher software version 4.10.1 (Gene Codes Corporation, Ann Arbor, MI). KDM6A was analyzed in samples with no MLL2 mutation using HRM analysis and Sanger sequencing as above (n = 37). For male samples, genotyping using spike-in control male genomic DNA (10%) was performed to detect a hemizygous mutation. The latter 23 patients (KMS-70 to KMS-92), as well as 22 patients with no mutation in either gene detected by conventional methods, were analyzed by targeted resequencing as described in the following section. We judged a variant as pathogenic when it was previously reported to cause KS, or novel variant when it was not observed in unaffected parents or in in-house exome data (n = 977), dbSNP135, or EVS6500 (Exome Variant Server, NHLBI GO Exome Sequencing Project, Seattle, WA; http://evs.gs.washington.edu/EVS/; accessed March 1, 2013). In addition, the missense mutation predicted to be polymorphism by both of two predictions (Polyphen-2: http://genetics.bwh.harvard.edu/pph2/ [Adzhubei et al., 2010] and MutationTaster: http://www.mutationtaster.org/ [Schwarz et al., 2010]) was considered to be non-pathogenic. Parentage analysis was conducted for the patients only when the parental samples were available. TaKaRa Ex Taq and TaKaRa LA Taq (both Takara, Tokyo, Japan) were used for amplification. The primer sequences and PCR conditions are available on request. All pathological variants were confirmed by Sanger sequencing. Nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence (RefSeq NM_003482.3 for MLL2, RefSeq NM_021140.2 for KDM6A).
Targeted Resequencing of MLL2 and KDM6A by Next-Generation Sequencing
Ion AmpliSeq Custom Panels (Life Technologies, Inc., Grand Island, NY) covering the entire coding region of MLL2 and KDM6A were created via the Ion AmpliSeq Designer v1.2 (https://ampliseq.com/browse.action). Libraries were prepared using the Ion AmpliSeq Library Kit 2.0 (Life Technologies, Inc.), with 10 ng of genomic DNA for each primer pool (two pools for this analysis). An Agilent 2200 TapeStation (Agilent Technologies, Santa Clara, CA) and the associated High Sensitivity D1K Screen Tape (Agilent Technologies) were used to check the size distribution and the concentration of the DNA libraries. Emulsion PCR and enrichment steps were carried out using the Ion OneTouch 200 Template Kit v2 (Life Technologies, Inc.). The amplicon libraries were sequenced on an Ion Torrent Personal Genome Machine system using 314 or 316 chips, and bar-coding was applied with an Ion Xpress Barcode Adapters 1–16 Kit (all Life Technologies, Inc.). Torrent Suite 2.2 (Life Technologies, Inc.) was used for mapping, base calling, and variant calling. Sequences were annotated using SeattleSeq Annotation 134 (http://snp.gs.washington.edu/SeattleSeqAnnotation134/). All variants were confirmed by Sanger sequencing.
Whole Exome Sequencing by High-Throughput Next-Generation Sequencing
Whole exome sequencing was performed in five individuals (KMS-09, -18, -21, -23, and -61) who had no MLL2 or KDM6A abnormality by HRM analysis. DNA was processed with a SureSelect Human All Exon V4 kit (Agilent Technologies), sequenced on a HiSeq2000 (Illumina, Inc., San Diego, CA), and analyzed as previously described [Tsurusaki et al., 2013]. Variants in MLL2 and KDM6A were confirmed by Sanger sequencing.
X-Inactivation Assay
X-inactivation analysis was performed as described [Allen et al., 1992] with slight modification. Briefly, genomic DNA (500 ng) was digested with two methylation-sensitive restriction enzymes, HpaII and HhaI (New England Biolabs, Beverly, MA), and purified by phenol/chloroform extraction and ethanol precipitation. Digested and undigested DNA samples (10 ng) were separately amplified for the (CAG)n polymorphism at the androgen receptor locus. The forward primer was labeled with 5′ FAM dye. PCR products were analyzed on an ABI 3500xl Genetic Analyzer using GeneMapper Software Version 4.1 (Applied Biosystems). The assay was independently performed twice.
cDNA Sequencing
Total RNA was extracted from a lymphoblastoid cell line established from KMS-81 (c.1909_1912del in KDM6A) using an RNeasy Plus mini kit (Qiagen, Hilden, Germany) with and without cycloheximide treatment (30 µg/ml) for 4 hr before cell collection. Reverse transcription (RT) was performed using a Superscript III First-Strand synthesis system for RT-PCR (Life Technologies, Inc.). As the mutation was located in exon 17, the region from the exon 15/16 boundary to the exon 17/18 boundary of KDM6A was amplified using cDNA-specific primer pairs (sequences available on request) and sequenced by the Sanger method.
Statistical Analysis
The frequencies of clinical features in the two groups were compared by Fisher's exact test. A difference was considered statistically significant if P < 0.05. Correction for multiple testing was not applied.
RESULTS
Overall Mutation Detection Rates
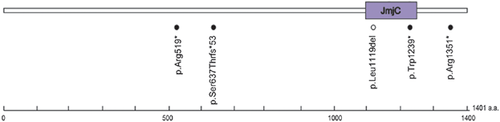
Pathogenic mutations in MLL2 and KDM6A were found in 50 (61.7%) and five (6.2%) of the 81 patients with KS, respectively (Figs. 1 and 2, Tables I and II). Of the 50 MLL2 mutations, 35 (70.0%) were predicted to be protein truncating-type and 15 (30.0%) were predicted to be non-truncating-type. Interestingly, non-truncating mutations were mostly localized in or adjacent to the functional domains, while truncating mutations were scattered throughout the entire coding region (Fig. 1). Fifteen of the MLL2 mutations have been previously reported (Table I). Three novel variants (not included in the 50 mutations) were considered non-pathogenic (Supplemental Table I). Variant c.10942C > G in patient KMS-22 was predicted to be benign by Polyphen-2 and MutationTaster, c.8813C > T in patient KMS-62 was inherited from an unaffected father, and c.4065A > T in KMS-75 was found heterozygously in our 977 in-house controls. An in-frame duplication in patients KMS-40 and KMS-62, which predicted p.Asp5055_Leu5056dup, was predicted to be polymorphic by MutationTaster, but was previously reported as a pathogenic mutation [Micale et al., 2011]. In addition, the other in-frame mutation in KMS-02 was also predicted to be polymorphic. Unfortunately, parental samples were unavailable for these individuals, except for the mother of patient KMS-62, who did not have this mutation; thus, the de novo status remains unclear. Of the five KDM6A mutations including three mutations reported previously [Miyake et al., 2013], four were truncating-type and one was an in-frame deletion located within the Jumonji C domain (Fig. 2).
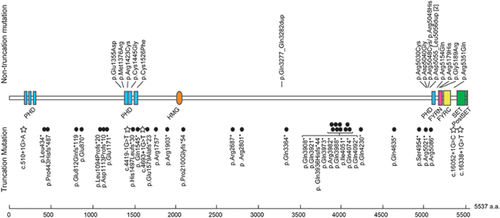
| Patient ID | Method | Mutation | Predicted amino acid change | De novo | Remarksa |
|---|---|---|---|---|---|
| Patients with MLL2 mutations | |||||
| KMS-02 | H | c.9831_9848dup | p.Gln3277_Gln3282dup | Unknown | Novel |
| KMS-08 | H | c.12688C > T | p.Gln4230* | Yes | Hannibal et al. [2011] |
| KMS-13 | H | c.2433_2434insCA | p.Glu812Glnfs*119 | Yes | Novel |
| KMS-14 | H | c.11806_11807dup | p. Gln3936Hisfs*44 | Yes | Novel |
| KMS-15 | H | c.15119A > G | p.Asp5040Gly | Yes | Novel |
| KMS-17 | H | c.5707C > T | p.Arg1903* | Yes | Novel |
| KMS-18 | W | c.12151delA | p.Ile4051* | Yes | Novel |
| KMS-20 | H | c.1300delC | p.Leu434* | Unknown | Novel |
| KMS-21 | W | c.3326_3336dup | p.Asp1113Profs*10 | Unknown | Novel |
| KMS-22 | H | c.4127T > G | p.Met1376Arg | Unknown | Novel |
| KMS-23 | W | c.15461G > A | p.Arg5154Gln | Unknown | Li et al. [2011] |
| KMS-24 | H | c.2608 G > T | p.Glu870* | Unknown | Novel |
| KMS-25 | H | c.11917C > T | p.Gln3973* | Unknown | Novel |
| KMS-27 | H | c.15142C > T | p.Arg5048Cys | Yes | Hannibal et al. [2011], Makrythanasis et al. [2013] |
| KMS-28 | H | c.14861C > A | p.Ser4954* | Unknown | Novel |
| KMS-29 | H | c.4419-1G > T | splice site | Unknown | Novel |
| KMS-30 | H | c.4633C > T | p. Gln1545* | Unknown | Novel |
| KMS-32 | H | c.8059C > T | p.Arg2687* | Unknown | Banka et al. [2012b] |
| KMS-33 | T | c.11962C > T | p.Gln3988* | Unknown | Novel |
| KMS-36 | H | c.4736_4737delAG | p.Glu1579Alafs*23 | Unknown | Novel |
| KMS-38 | H | c.15143G > A | p.Arg5048His | Unknown | Makrythanasis et al. [2013] |
| KMS-40 | T | c.15163_15168dup | p.Asp5055_Leu5056dup | Unknown | Micale et al. [2011] |
| KMS-41 | H | c.1328delC | p.Pro443Hisfs*487 | Yes | Ng et al. [2010] |
| KMS-42 | H | c.16052G > A | p.Arg5351Gln | Yes | Novel |
| KMS-43 | H | c.510 + 1G > A | splice site | Unknown | Novel |
| KMS-49 | T | c.15565G > A | p.Gly5189Arg | Unknown | Novel |
| KMS-51b | H | c.6297_6298delAC | p.Pro2100Glyfs*54 | Yes | Novel |
| KMS-52 | H | c.4693 + 1G > T | splice site | Yes | Novel |
| KMS-53 | H | c.10090C > T | p.Gln3364* | Yes | Novel |
| KMS-54 | T | c.8401C > T | p.Arg2801* | Yes | Novel |
| KMS-56 | H | c.15536G > A | p.Arg5179His | Yes | Ng et al. [2010] |
| KMS-58 | H | c.4333T > G | p.Cys1445Gly | Yes | Novel |
| KMS-59 | H | c.15256C > T | p.Arg5086* | Yes | Banka et al. [2012b] |
| KMS-60 | H | c.11761C > T | p.Gln3921* | Unknown | Novel |
| KMS-61c | W | c.5269C > T | p.Arg1757* | Yes | Novel |
| KMS-62 | H | c.15163_15168dup | p.Asp5055_Leu5056dup | Unknownf | Micale et al. [2011] |
| KMS-63 | T | c.4577G > T | p.Cys1526Phe | Yes | Novel |
| KMS-69 | H | c.11944C > T | p.Arg3982* | Yes | Paulussen et al. [2011] |
| KMS-70 | T | c.13903C > T | p.Gln4635* | Yes | Novel |
| KMS-71d | T | c.12220C > T | p.Gln4074* | Unknown | Novel |
| KMS-72 | T | c.15061C > T | p.Arg5021* | Yes | Banka et al. [2012b] |
| KMS-73 | T | c.12274C > T | p.Gln4092* | Unknown | Micale et al. [2011] |
| KMS-76 | T | c.4490_4491delAC | p.His1497Leufs*30 | Yes | Novel |
| KMS-78 | T | c.16338 + 1G > T | splice site | Yes | Novel |
| KMS-80 | T | c.15088C > T | p.Arg5030Cys | Unknown | Makrythanasis et al. [2013] |
| KMS-82 | T | c.3511G > T | p.Glu1171* | Yes | Novel |
| KMS-85 | T | c.11722C > T | p.Gln3908* | Yes | Paulussen et al. [2011] |
| KMS-87 | T | c.3281_3282delTC | p.Leu1094Profs*20 | Yes | Novel |
| KMS-88 | T | c.16052 + 1G > C | splice site | Unknown | Novel |
| KMS-91 | T | c.4267C > T | p.Arg1423Cys | Unknown | Novel |
| Patients with KDM6A mutations | |||||
| KMS-31e | H | c.3717G > A | p.Trp1239* | Unknown | Miyake et al. [2013] |
| KMS-37e | H | c.1555C > T | p.Arg519* | Unknown | Miyake et al. [2013] |
| KMS-65e | H | c.3354_3356delTCT | p.Leu1119del | Yes | Miyake et al. [2013] |
| KMS-81 | T | c.1909_1912delTCTA | p.Ser637Thrfs*53 | Yes | Novel |
| KMS-83 | T | c.4051C > T | p.Arg1351* | Unknown | Novel |
- H, high-resolution melting analysis/Sanger sequencing; T, targeted resequencing; W, whole exome sequencing.
- RefSeq NM_003482.3 for MLL2 and RefSeq NM_021140.2 for KDM6A were used as reference sequences.
- a References are listed when the same mutation has been reported previously.
- b This patient was reported as proband 1 by Tekin et al. [2006].
- c The detailed clinical features of this patient were reported by Ito et al. [2013] because of her hypothalamic pituitary complications.
- d The clinical course of this patient, particularly the idiopathic thrombocytopenic purpura, was reported by Torii et al. [2009].
- e These patients have been reported in our previous study [Miyake et al., 2013].
- f Patient KMS-62: no mutation in the mother.
| Amino acid changea | Patient ID | Domain | Polyphen-2 (score) | MutationTaster |
|---|---|---|---|---|
| p.Met1376Arg | KMS-22 | — | Probably damaging (0.915) | Polymorphism |
| p.Arg1423Cys | KMS-91 | PHD | Probably damaging (1.000) | Disease causing |
| p.Cys1445Gly | KMS-58 | PHD | Probably damaging (1.000) | Disease causing |
| p.Cys1526Phe | KMS-63 | PHD | Probably damaging (0.999) | Disease causing |
| p.Gln3277_Gln3282dup | KMS-02 | — | NA | Polymorphism |
| p.Arg5030Cys | KMS-80 | — | Probably damaging (1.000) | Disease causing |
| p.Asp5040Gly | KMS-15 | — | Probably damaging (1.000) | Disease causing |
| p.Arg5048Cys | KMS-27 | — | Probably damaging (1.000) | Disease causing |
| p.Arg5048His | KMS-38 | — | Probably damaging (1.000) | Disease causing |
| p.Asp5055_Leu5056dup | KMS-40, 62 | — | NA | Polymorphism |
| p.Arg5154Gln | KMS-23 | — | Probably damaging (1.000) | Disease causing |
| p.Arg5179His | KMS-56 | FYRN | Possibly damaging (0.840) | Disease causing |
| p.Gly5189Arg | KMS-49 | FYRN | Probably damaging (1.000) | Disease causing |
| p.Arg5351Gln | KMS-42 | — | Probably damaging (1.000) | Disease causing |
- a The nucleotide mutation nomenclature for these predicted protein mutations are included in Table I.
Clinical Comparison Between the Mutation-Positive and -Negative Groups
We compared the clinical features of the MLL2 or KDM6A mutation-positive and -negative groups (Supplemental Table II). Long palpebral fissures were observed in almost all patients. Cleft lip/palate was more frequently observed in the mutation-positive group (P = 0.0197). Interestingly, developmental delay and intellectual disability were observed in all individuals with mutations but were unobserved in some mutation-negative cases (P = 0.0314 and P = 0.1778, respectively). Blue sclera, lower lip pits, spine/rib abnormality, hip joint dislocation, umbilical hernia, kidney dysfunction, cryptorchidism, liver abnormality, spleen abnormality, premature thelarche, neonatal hyperbilirubinemia, and anemia were observed only in the mutation-positive group.
Clinical Comparison of the MLL2-Mutated and KDM6A-Mutated Groups
We compared the clinical features between the MLL2-mutated and KDM6A-mutated groups (Figs. 3-5, Supplemental Table III). High arched eyebrows, short fifth fingers, and hypotonia in infancy were more frequent in individuals with MLL2 mutations than in individuals with KDM6A mutations (P = 0.0364, 0.0039, and 0.0283, respectively). Short stature was more frequent in individuals with KDM6A mutations (P = 0.0485). Although not statistically significant, postnatal growth retardation was observed in all individuals with KDM6A mutations, whereas this was observed in only half of the individuals with MLL2 mutations.
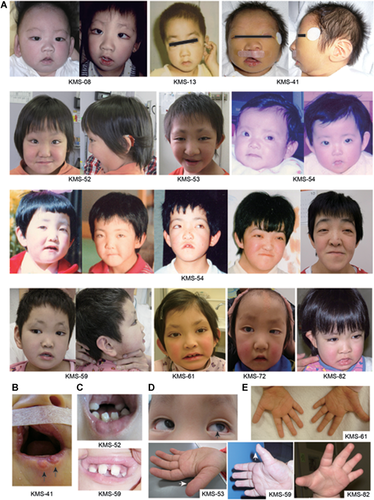
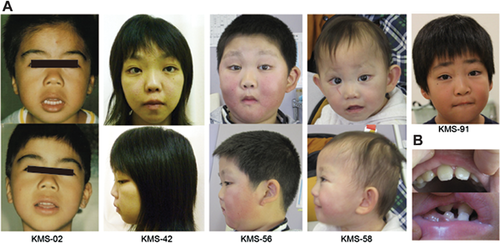
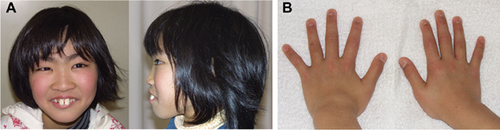
Clinical Comparison of Individuals With a MLL2 Truncating-Type and Non-Truncating-Type Mutation
Most clinical features were observed at a similar ratio in both groups (Supplemental Table IV), except for prominent ears and hypotonia, which were more frequently observed in the truncating-type group than in the non-truncating-type group (P = 0.0339 and P = 0.0248, respectively). However, the facial appearance of individuals in the truncating-type group was more typical, based on the ten originally reported patients with KS [Kuroki et al., 1981; Niikawa et al., 1981], than that in the non-truncating-type group (Figs. 3 and 4). Except for patient KMS-58, the facial appearance of patients with a non-truncating-type mutation was rather less typical. It should be noted that these patients had thick eyebrows (not present in patient KMS-56). Furthermore, ectropion of the lower eyelid, depressed nasal tip, short columella, and prominent ears all seemed less obvious in the individuals with a non-truncating-type mutation.
X-Inactivation Pattern in Female Patients With a KDM6A Mutation
A KDM6A mutation was identified in two females (KMS-65 and KMS-81). Individual KMS-65 (c.3354_3356del, which predicts p.Leu1119del) showed a random X-inactivation pattern [Miyake et al., 2013], while individual KMS-81 with a frame-shift mutation showed marked skewing (98:2; Supplemental Fig. 1A). By RT-PCR using mRNA derived from a lymphoblastoid cell line from patient KMS-81, we confirmed that both the mutated and normal alleles were transcribed at similar levels when nonsense-mediated mRNA decay (NMD) was inhibited by cycloheximide treatment (Supplemental Fig. 1B). In untreated cells, or cells treated with dimethylsulfoxide (negative control), the mutant allele was transcribed at a lower level than the wild-type allele, indicating that NMD partially eliminated the mutant.
Exome Sequencing
Among the five patients who were also analyzed by whole exome sequencing, mutations were identified and later confirmed by Sanger sequencing in four (Table I). Fragments with an 11 base-pair insertion (c.3326_3336dup) of MLL2 in patient KMS-21 could not be amplified by Ex Taq, but could be amplified by LA Taq with LA buffer and confirmed by Sanger sequencing. Three other mutations were missed by HRM analysis (Table I).
DISCUSSION
We identified 50 MLL2 and five KDM6A mutations among 81 patients with KS and add to the 246 MLL2 mutations described in patients with KS [Ng et al., 2010; Hannibal et al., 2011; Li et al., 2011; Micale et al., 2011; Paulussen et al., 2011; Banka et al., 2012a, 2012b; Kokitsu-Nakata et al., 2012; Tanaka et al., 2012; Bogershausen and Wollnik, 2013; Makrythanasis et al., 2013] (Human Gene Mutation Database Professional 2012.3; https://portal.biobase-international.com/hgmd/pro/gene.php). Our mutation-positivity rate for either gene was 67.9% (55/81), and that for MLL2 only was 61.7% (50/81); these figures are compatible with those reported in a review (55–80%) [Banka et al., 2012b]. Mutation-negative patients suggest the existence of unknown genes to cause KS or misdiagnosis.
As for the phenotype–genotype relationship, Banka et al. [2012b] suggested that feeding problems, kidney anomalies, premature thelarche, joint dislocation, and palatal malformation were more frequently observed in patients with MLL2-mutations than in patients with normal MLL2 sequence. Hannibal et al. [2011] reported that renal anomalies were more common in patients who had MLL2 mutations compared to those who did not. Li et al. [2011] reported that short stature and renal anomalies were more frequent in patients with MLL2-mutations than in those with normal MLL2 sequence. In our study, premature thelarche was observed only in patients with MLL2 mutations, but this was not significant (P = 0.1137). The frequencies of kidney anomalies, hip joint dislocation, and short stature were not different when comparing those with and without MLL2 mutations (P = 0.3030, P = 1.0000, and P = 0.0717, respectively; Supplemental Table V). High arched eyebrows, palatal malformation (cleft palate/lip), low posterior hairline, and short fifth finger were more frequently observed in individuals with MLL2 mutations than in patients with normal MLL2 (P = 0.0118, P = 0.0284, P = 0.0493, and P = 0.0137, respectively; Supplemental Table V).
X-inactivation skewing in patients with KS has been discussed since the discovery of the KDM6A deletion in a female with KS [Lederer et al., 2012; Miyake et al., 2013]. In two female patients reported here, patient KMS-65, who had an in-frame deletion, showed a random X-inactivation pattern, but patient KMS-81, who had a truncating-type mutation, showed marked skewing. X-inactivation skewing was also reported in two affected females with KDM6A deletion reported by Lederer et al. [2012]. cDNA sequence analysis of patient KMS-81 indicated that the mutant allele of KDM6A was expressed at a similar level to the wild-type allele under NMD inhibition. This result suggests that KDM6A mostly escapes X-inactivation in female lymphoblastoid cells. Interestingly, KDM6A/Kdm6a escapes X-inactivation in humans and mice, and in mice its expression level from the inactive X chromosome (Xi) was reported as 15–35% of that from the active X chromosome (Xa) [Greenfield et al., 1998; Xu et al., 2008]. We calculated the hypothetical expression assuming a 30% KDM6A expression level from Xi and 100% expression from Xa (Supplemental Fig. 2). In patient KMS-81, who showed marked skewing (98:2), either the mutant X chromosome or the wild-type X was inactivated in 98% of cells. If the mutant were inactivated, the expression level would be below 1 (1.0 × 0.98 + 0.3 × 0.02 = 0.986). If the wild-type were inactivated, the expression level would also be below 1 (1.0 × 0.02 + 0.3 × 0.98 = 0.314). The KS phenotype is usually unassociated with Turner syndrome (45,X), with the KDM6A expression level at 1.0 [Miyake et al., 2013]. It is possible that having a KDM6A expression level of 1.0 is essential for a normal human phenotype. Similarly, males with only one copy of KDM6A do not manifest KS. We previously mentioned the possibility of UTY compensation for KMD6A (Supplemental Fig. 2) [Miyake et al., 2013], although human UTY lacks demethylase activity [Hong et al., 2007; Lan et al., 2007]. The recent evidence that XUtx−XUtx− homozygous mice demonstrated a more severe phenotype than XUtx−YUty+ mice indicates that UTY can compensate for the loss of UTX in embryonic development [Shpargel et al., 2012]. Because mouse and human UTY show 75% identity, and 95% identity in the Jumonji C domain [Shpargel et al., 2012], it is likely that normal human males who have only one copy of KDM6A are supplemented by UTY in a demethylase-independent manner.
Interestingly, XUtx−YUty+ mice showed small body size [Shpargel et al., 2012]. Similarly, the human KDM6A-mutated group exhibited short stature and postnatal growth retardation.
Regarding our mutation detection methods, HRM analysis and Sanger sequencing are both imperfect. Next-generation sequencing is more sensitive (especially for single nucleotide variants and small insertions/deletions), faster, and cheaper due to multiple gene screening and the potential to multiplex. However, a microdeletion involving MLL2 or KDM6A or low-level mosaicism of a single nucleotide variant might be missed by this method. Therefore, in patients who test mutation-negative, more comprehensive approaches might be necessary. In conclusion, we investigated MLL2 and KDM6A mutations and their clinical consequences in patients with KS. The majority of the clinical features were observed at a similar frequency among patients with either MLL2 or KDM6A-mutations. The genetic basis of the patients who tested mutation-negative (20–45%) remains elusive. Further studies are necessary to understand the whole picture of the genetic aspects of KS and its genotype–phenotype relationships.
ACKNOWLEDGMENTS
We thank the patients and their parents for participating in this work. We also thank Ms. Y. Yamashita, Ms. E. Koike, Ms. S. Sugimoto, Ms. N. Watanabe, Ms. K. Takabe, and Mr. T. Miyama for their technical assistance. This work was supported by research grants from the Ministry of Health, Labour and Welfare of Japan (H. Saitsu, N. Matsumoto, N. Miyake), the Japan Science and Technology Agency (N. Matsumoto), the Strategic Research Program for Brain Sciences (N. Matsumoto), a Grant-in-Aid for Scientific Research on Innovative Areas-(Transcription cycle)-from the Ministry of Education, Culture, Sports, Science and Technology of Japan (N. Matsumoto), a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (N. Matsumoto), a Grant-in-Aid for Young Scientists from the Japan Society for the Promotion of Science (H.S., N. Miyake), the Takeda Science Foundation (N. Matsumoto, N. Miyake), the Yokohama Foundation for the Advancement of Medical Science (N. Miyake), and the Hayashi Memorial Foundation for Female Natural Scientists (N. Miyake).